QnrVC, was described in 2008, within an integron environment in Vibrio cholerae.1 Further studies have described QnrVC-like sequences, either in V. cholerae or other microorganisms, located within integrons or plasmids, some of them able to be transferred within microorganisms.2–7
This work analyses the phylogenetic relationships of QnrVC respecting the 5 established plasmid-encoded Qnr-families, searching GenBank for QnrVC-related sequences.8
A GenBank search was performed using the nucleotide (GenBank: EU436855) and amino acid (GenBank: ACC54440.2) sequences of QnrVC1/VC3 as template, selecting those sequences with identities levels higher than 70% with respect to QnrVC1/VC3.8
Those selected sequences and representative sequences of the established Qnr-families present in http://www.lahey.org/qnrStudies, were included in the phylogenetic studies.
The BIOEDIT software was used to perform an in silico alignment to establish the presence of the characteristic loops A and B structures.9
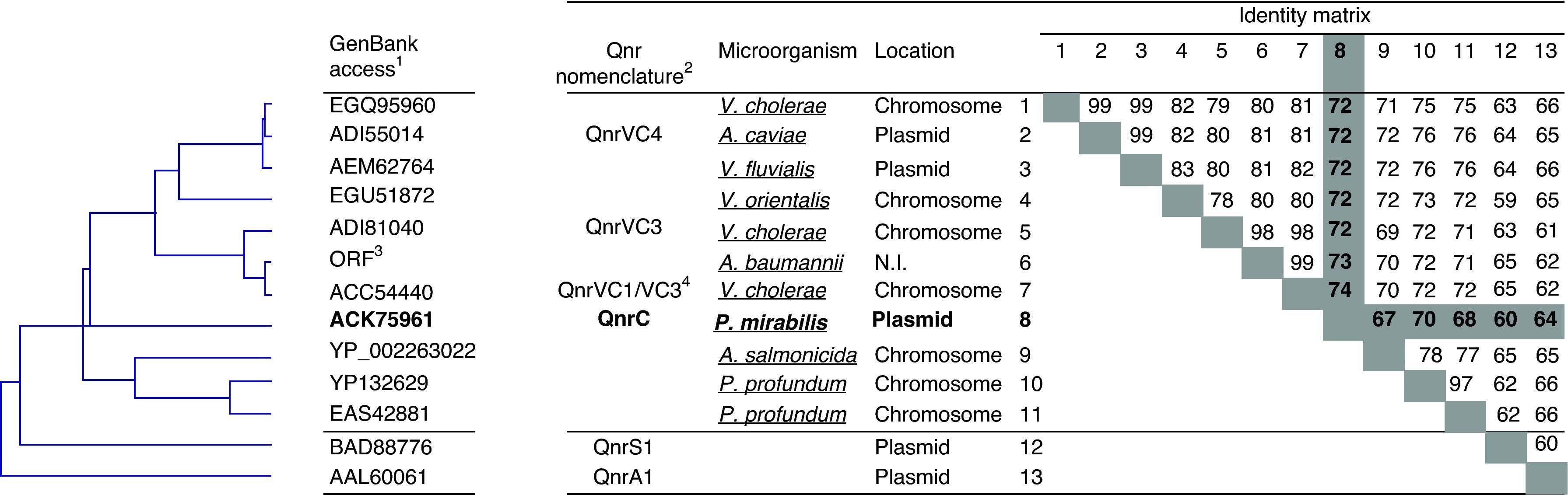
Ten different Qnr sequences, which following established criteria8 may be classified within a common family together QnrVC1/VC3, were found in GenBank (Fig. 1). Thus, 6 chromosomal-encoded sequences (ADI81040 and EGQ95960 from V. cholerae; YP132629 and EAS42881 from Photobacterium profundum; EGU51872 or EEX92304 from Vibrio orientalis; YP_002263022 from Allivibrio salmonicida: 3 plasmid-encoded (1 from Aeromonas caviae – ADI55014; 1 from Vibrio fluvialis – found under 3 different GenBank entries: AEM62764, AER42862 or AER42863; and QnrC) and an ORF from Acinetobacter baumannii (DNA GenBank: GU944730), without information about its chromosomal or plasmid encoding, were detected. This ORF, found within a class 1 integron, shows the transferability of QnrVC-like from water-borne microorganisms towards a relevant nosocomial pathogen.6 Besides, sequences with internal stop codons were also found, as QnrVC2 (GenBank: AB200915) present in the pVN84 of V. cholerae.1
Phylogenetic relationships among qnrVC-like genes. The distance-based tree was generated by using p distance with the neighbour-joining method. 1: only one GenBank access is indicated for each different Qnr sequence included in the figure, as indicated in the text, QnrVC4 has also been described in other microorganisms. 2: specific nomenclature present in the literature. When the sequence has been published or introduced in Genbank as an erroneous member of an established Qnr family has not been considered. 3: located within the GenBank DNA sequence No. GU944730. 4: introduced in GenBank as QnrVC1 in 2008,1 but a posterior correction in the sequence results in a sequence 100% identical to GenBank sequences ADI81036 and ADI81043 introduced as QnrVC3.3 In the absence of an established nomenclature we refer to this sequence as QnrVC1/VC3. 5: YP_002263022 (A. salmonicida) has been claimed as a representative of a novel Qnr-family, however it is not plasmid encoded and many other QnrVC-like were previously described, then we considered that will be more correct considered as a potential member of the QnrVC family. N.I.: non-information about its genetic location is disposable. In the identity matrix, QnrC has been noted in bold, and identities of QnrC with remaining sequences have been highlighted.
The plasmid-encoded QnrVC-like (QnrVC4) present in A. caviae have also been recently introduced in GenBank as detected in river-isolated microorganisms such as Escherichia coli (JQ837999.1), A. hydrophyla (i.e.: JQ838001.1), in both encoded within a class 1 integron, Pseudomonas sp. (i.e.: JQ838008.1), or other Aeromonas spp. (JQ838004.1). QnrC shows identities >70% with the remaining potential QnrVC sequences, except EAS42881 (P. profundum) and YP_002263022 (A. salmonicida) (Fig. 1). This phylogenetic nearby between QnrC and members of the QnrVC family, previously noted by Wang et al.,10 suggests the Vibrionaceae genus as the ancestral chromosomal source of QnrC.
A serious confusion statement was observed among GenBank entries. Thus, 5 entries, in which a QnrVC-like was found, were recorded as QnrB1, other 2 as QnrC (both different that currently described QnrC), and two different sequences as QnrVC3. Similarly, the ORF present in sequence GU944730, it is not reported in GenBank as encoding a QnrVC-like gene, but it has been introduced in INTEGRALL (http://integrall.bio.ua.pt/), an internet repository devoted to integrons, as encoding a QnrVC1b protein, and recently published as QnrVC-like.6
Finally, the Qnr loops A and B, essential for the activity of different Qnr proteins,9 were predicted in all QnrVC-related proteins. Thus, QnrVC-like may be considered as full functional Qnr-like proteins.
In conclusion, a series of sequences that may be including within a new common transferable Qnr-family have been found in GenBank. The close similarity (higher than 70%) between the QnrC and QnrVC families may suggest the need for nomenclature unification following the current established normative.
Joaquim Ruiz has a fellowship from the Programa I3 from Ministerio de Economía y Competitividad, Spain.