Previous knowledge of molecular mechanisms related with multi-drug resistances in tuberculosis is important if molecular diagnostic procedures want to be used in specific geographical regions. For that reason, the aim of this study was to investigate the mutations at rpoB, katG and inhA in multi-drug resistant tuberculosis isolates from Southeast Mexico.
MethodsIsolates of tuberculosis with a confirmed resistance against rifampicin and isoniazid were collected and sequencing analysis was performed of the rpoB rifampicin resistance-determining region, the katG and the encoding region of inhA.
ResultOf 74 isolates with multidrug resistance, 34 (46%) presented six mutations in katG; the most abundant was katG315 in 29 (39%) isolates. At inhA, nine (11%) isolates presented three mutations; the most frequent was inhA21, located in five (6%) strains. Eleven polymorphisms were observed at rpoB in 61 (82%) isolates, prevailing rpoB531 and rpoB 526 in 48 (64%) and ten (12%) isolates, respectively. Eleven double combinations were observed in 39 (52%) isolates, the most common of which was rpoB531+katG315, found in 22 (29%) strains.
ConclusionThis study provides valuable information on the diversity of polymorphisms in genes related to multidrug-resistant tuberculosis, as well as the presence of new mutations not previously described; this information should be considered in the implementation of molecular diagnostic tests.
El conocimiento previo de los mecanismos moleculares relacionados con las multi-farmacorresistencia es de suma importancia si se desean emplear procedimientos de diagnóstico molecular en regiones geográficas específicas. Por esa razón, el objetivo de este estudio fue investigar las mutaciones en rpoB, katG e inhA en cepas aisladas de tuberculosis multirresistente circulantes en el sureste de México.
MétodosSe recuperaron aislados de tuberculosis con una resistencia confirmada a rifampicina e isoniazida, y se realizó la secuenciación y posterior análisis de la región determinante de la resistencia a rifampicina en el gen rpoB, así como del gen katG y la región codificante de inhA.
ResultadoDe 74 cepas con multirresistencia, solo 34 (46%) presentaron 6 mutaciones en katG; la más abundante fue katG315 presente en 29 (39%) aislados. En inhA, solo 9 (11%) aislados presentaron 3 mutaciones; la más frecuente fue inhA21, localizada en 5 (6%) cepas. Se observaron en 61 (82%) cepas 11 polimorfismos en rpoB, siendo los más frecuentes rpoB531 en 48 (64%) y rpoB526 en 10 (12%) aislados, respectivamente. Se observaron 11 combinaciones dobles en 39 (52%) cepas, la más común fue rpoB531+katG315, presente en 22 (29%) cepas.
ConclusiónEste estudio proporciona información valiosa sobre la diversidad de polimorfismos en genes relacionados con la tuberculosis multirresistentes, así como la presencia de nuevas mutaciones no descritas previamente, esta información deberá ser considerar en la implementación de pruebas de diagnóstico molecular.
The 2017 global tuberculosis report of the World Health Organization (WHO) estimated that, in 2016, there were 10.5 million new cases and 1.5 million deaths as a result of Tuberculosis.1 Lack of adequate treatment, often due to irregular drug supply, mismanagement of patients, inadequate administration of antimicrobial therapy regimens and poor patient compliance are the most important factors in the emergence of drug resistant tuberculosis (DR-TB). According to WHO, 3 to 5% of DR-TB cases exhibit simultaneous resistance to the most important first line drugs isoniazid and rifampicin, known as multi-drug-resistant tuberculosis (MDR-TB). At present, DR-TB and MDR-TB are a major public health concern in several countries, complicating the epidemiological landscape and control of this disease, and creating a serious concern for global public health.2
Several studies have demonstrated that 85–90% of isolates with resistance against rifampicin had a mutation in an 81-bp region of the rpoB, this gene has an extension of 3543pb and codify to the B sub-unit of the RNA polymerase.3–6 Meanwhile, 30–70% of strains with resistance toward isoniazid had mutations at the katG and inhA genes. katG has a long of 2223pb and synthesizes a catalase-peroxidase enzyme involved in the synthesis of mycolic acid, while inhA and its regulator give rise to inhA protein, involved in the synthesis of fatty acids.3,6–9 However, the percentages of mutations associated with these genes show variations according to geographical areas, with consequent implications for the implementation of molecular MDR-TB detection methods.4,6,7,10
Mexico occupies third place for incidence of TB and MDR-TB in Latin America. However, reports providing characterization of the mutations associated with rpoB, katG and inhA in MDR-TB isolates are limited.11–15 The State of Veracruz contributes almost 15% of the 20,000 cases reported in the country annually, and is placed among the five states nationally (Veracruz, Nuevo Leon, Tamaulipas, Guerrero and Baja California) with significant numbers of aggravated forms of TB, such as poly-, multi- and, more recently, extreme-drug resistance.16 The objective of this study is therefore to characterize the mutations associated with rpoB, katG and inhA genes in multi-drug resistant clinical isolates of M. tuberculosis from Veracruz, Mexico, and to discuss the usefulness of these mutations in the context of actual molecular diagnostic procedures.
MethodsCollection of clinical samples, isolation of mycobacteria and drug susceptibility testFrom 2007 to 2012, M. tuberculosis strains from Veracruz, Mexico, were randomly recovered from sputum samples of patients with clinical manifestations compatible with drug resistance (Individuals who, despite receiving treatment, maintain a productive cough as well as the presence of bacilli in the sputum or other specimens at the end of treatment). Definition for a new case and undergone re-treatment were according to the Mexican official standard (http://dof.gob.mx/nota_detalle.php?codigo=5321934&fecha=13/11/2013). Sputum decontamination was performed using N-acetyl-l-Cysteine-NaOH and primary isolation of mycobacteria was developed with Löwenstein–Jensen medium. Susceptibility testing against the first line drugs Streptomycin (S), Isoniazid (H), Rifampicin (R), Ethambutol (E) and Pyrazinamide (Z) was conducted using the fluorometric method (MGIT, Becton-Dickinson, MD). Social and clinical variables of individuals included in the study were recovered from the clinical summary.
No physical interventions occurred with the patients, and all information collected was confidential. Ethical issues derived from this study were overseen by the respective committee of the Public Health Institute of the University of Veracruz.
DNA purification and PCR-amplification of rpoB, katG and inhAA loop of fresh culture isolate was suspended in 500uL of TE buffer [10mM Tris/HCl (pH 8.0), 1nM EDTA] and DNA was further purified following Van Soolingen.17 The DNA concentration was determined in a Nanodrop 100 (ThermoScientific, USA) and the DNA stored at −20°C until subsequent use.
The following three loci were amplified using locus-specific primers: the well-established region associated with resistance to Isoniazid at katG: katG-F (5′-GCAGATGGGGCTGATCTACG-3′) and katG-R (5′-AACTCGTCGGCCAATTCCTC-3′) (Zenteno-Cuevas 2009), and a fragment of 517pb of inhA gene: inhA-F (5′-AGGTCGCCGGGGTGGTCAGC-3′) and inhA-R (5′-AGCGCCTTGGCCA TCGAAGCA-3′) (Morlock 2003). Finally, the rpoB gene including the rifampicin resistance-determining region (RRDR): rpoB-F (5′-AGCGGATGACCACCCAGGAC-3′) and rpoB-R (5′-TCAG GGGTTTCGATCGGGCA-3′) (Zenteno-Cuevas 2009).
PCR-amplification conditions and sequencing of amplicons were adapted considering to Zenteno et al. (2009). The PCR reaction mix contained: 10mM Tris pH 8, 1.5mM MgCl2, 0.2mM of each deoxynucleotide triphosphate, 10μM of forward and reverse primers, 1.25U Taq polymerase (Promega, USA), 5% glycerol, 100ng DNA template, and free nuclease water to reach a final volume of 25μl. Amplifications were performed individually for each gene in a Verity thermal Cycler (Applied Biosystems, USA), in all loci these were the cycling conditions: initial denaturation step of 95°C for 3min; 35 cycles of 95°C for 40s, 57°C for 30s and 72°C for 1min, and a final extension step of 72°C for 3min.
Products were electrophoretically separated in a 1.5% agarose gel, then purified using Amicon Ultra centrifugal filters (Millipore, Ireland). The final concentration was determined by electrophoresis, using the Mass Ruler Low Range DNA Ladder (Fermentas, USA).
DNA sequencing and sequence analysisThe sequencing reactions were performed in forward and reverse directions using 6μl of the Big Dye Terminator Cycle Sequencing Kit V3.1 (Applied Biosystems, USA), 3.2pM of PF and PR primers and 20ng of purified PCR product in a final volume of 20μl. The amplification conditions were 25 cycles of 95°C for 30s; 50°C for 15s and a final step of 60°C for 4min.
The products were purified using the ZR DNA sequencing clean-up kit™ (Zymo Research, USA), re-suspended in Hi-Di formamide (Applied Biosystems, USA), heated to 95°C for 5min, cooled on ice and finally loaded into a 96-well plate MicroAmp reaction plate (Applied Biosystems, USA).
The DNA products were sequenced by capillary electrophoresis in a Genetic Analyzer 3500 (Applied Bio-systems, USA). The fluorescence spectra were analyzed with the software Data Collection V1.01 (Applied Bio-systems, USA). Analysis of the sequences and identification of the mutations were performed using the sequencing Analysis V5.4 and SeqScape V2.7 programs (Applied Bio-systems, USA), respectively.
The wild type rpoB (Gene Bank accession number: 759807–763325), katG (Gene Bank accession number: 2153889–2156111) and inhA genes (Gene Bank accession number: 1673223–1675011) from M. tuberculosis H37Rv were used as reference sequences.
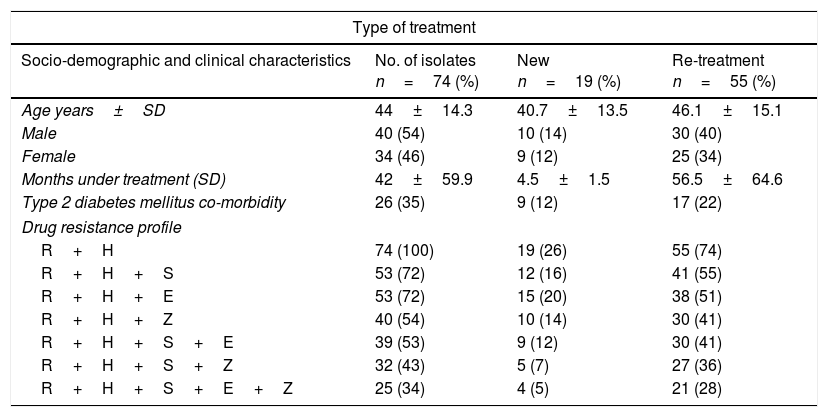
ResultsEpidemiological characteristics and drug sensitivity profilesOf the 352 drug resistant strains recovered, 74 (21%) presented simultaneous resistances against Isoniazid and Rifampicin (MDR-TB). Of the 74 individuals bearing a MDR-TB strain, mean age was 44±14.3 years and 54% (40/74) were male. The mean time under treatment for TB was 42±59 months. Use of alcohol and tobacco prior to diagnosis of TB was observed in 3% (2/74) and 16% (12/74) of the individuals respectively, and incidence of type 2 diabetes mellitus at the moment of diagnosis was found in 35% (27/74) of the individuals (Table 1). According to the type of treatment, 25% (19/74) of the individuals with MDR-TB were patients identified as a new case and 75% (55/74) had undergone re-treatment (Table 1).
Prevalence of drug resistance phenotypes among clinical multidrug resistant M. tuberculosis isolates (n=74).
| Type of treatment | |||
|---|---|---|---|
| Socio-demographic and clinical characteristics | No. of isolates n=74 (%) | New n=19 (%) | Re-treatment n=55 (%) |
| Age years±SD | 44±14.3 | 40.7±13.5 | 46.1±15.1 |
| Male | 40 (54) | 10 (14) | 30 (40) |
| Female | 34 (46) | 9 (12) | 25 (34) |
| Months under treatment (SD) | 42±59.9 | 4.5±1.5 | 56.5±64.6 |
| Type 2 diabetes mellitus co-morbidity | 26 (35) | 9 (12) | 17 (22) |
| Drug resistance profile | |||
| R+H | 74 (100) | 19 (26) | 55 (74) |
| R+H+S | 53 (72) | 12 (16) | 41 (55) |
| R+H+E | 53 (72) | 15 (20) | 38 (51) |
| R+H+Z | 40 (54) | 10 (14) | 30 (41) |
| R+H+S+E | 39 (53) | 9 (12) | 30 (41) |
| R+H+S+Z | 32 (43) | 5 (7) | 27 (36) |
| R+H+S+E+Z | 25 (34) | 4 (5) | 21 (28) |
Drug resistant combinations R+H+S and R+H+E were observed in 72% (53/74) individuals, of which 36 (27/74) were observed as new cases. Resistance to all first line drugs R+H+S+E+Z was found in 34% (25/74), of which 5% (4/74) were classified as new cases (Table 1).
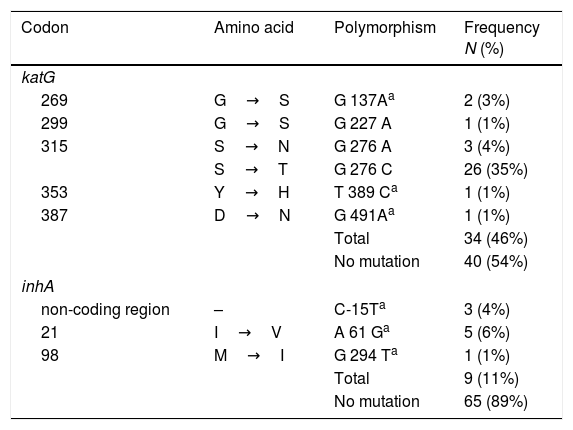
Mutations at inhA and katGThe analysis of the 555bp fragment of katG (from codon 224 to 408) and the 517pb fragment of inhA (from codon −115 to 134) showed that 46% (34/74) and 10% (8/74) of the isolates harbored a mutation within the katG and inhA genes, respectively (Table 2).
Mutations in the katG and inhA in MDR-TB isolates from Mexico, (n=74).
| Codon | Amino acid | Polymorphism | Frequency N (%) |
|---|---|---|---|
| katG | |||
| 269 | G→S | G 137Aa | 2 (3%) |
| 299 | G→S | G 227 A | 1 (1%) |
| 315 | S→N | G 276 A | 3 (4%) |
| S→T | G 276 C | 26 (35%) | |
| 353 | Y→H | T 389 Ca | 1 (1%) |
| 387 | D→N | G 491Aa | 1 (1%) |
| Total | 34 (46%) | ||
| No mutation | 40 (54%) | ||
| inhA | |||
| non-coding region | – | C-15Ta | 3 (4%) |
| 21 | I→V | A 61 Ga | 5 (6%) |
| 98 | M→I | G 294 Ta | 1 (1%) |
| Total | 9 (11%) | ||
| No mutation | 65 (89%) | ||
Six different polymorphisms were observed at katG; the most common was at S315T (276 G/C) found in 35% (26/74) of isolates, followed by S315N (G276A) in 4% (3/74) and G269S (G137A) in 3% (2/74). Three different changes were observed, each in single isolates, and no mutations were observed in 54% (40/74) of the isolates.
With respect to inhA, the most frequent polymorphism was found at I21V (A61G) in 6% (5/74) isolates, followed by mutation, in the non-coding region, C-15T in 4% (3/74). No mutations were observed in the remaining 89% (65/74) of the strains (Table 2).
Finally, is important to note that five mutations were described for the first time; katG269S, katGY353H, katGD387N, inhAI21V and inhAM98I; and 38% (20/74) of the isolates present no mutation in the katG and inhA genes.
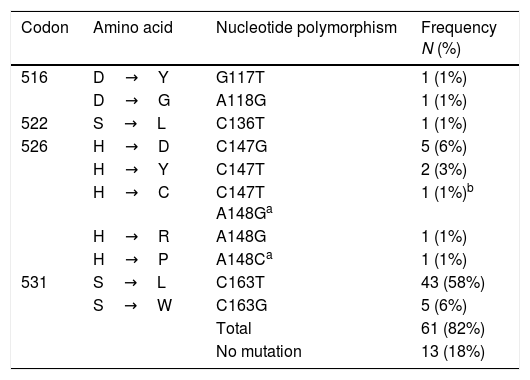
Mutations at rpoBTable 3 shows the polymorphisms found in the 266bp amplicon of the rpoB gene (from codon 477 to 565), considering the 81bp of the RRDR. A total of 82% (61/74) of the isolates harbored one mutation, and eleven polymorphisms were identified. The most common of these was S531L (C163T), found in 58% (43/74) of the isolates, followed by S531W (C163G) and H526D (C147G) in 6% (5/74), and H526Y (C147T) in 3% (2/74) of the strains, respectively. One isolate had a double mutation at codons 147C/T and 148 A/G, giving rise to the polymorphism H526C. This mutation 148 A/G at H526C together with A148C at H526P, are described for the first time.
Mutations in rpoB in MDR-TB isolates from Mexico (n=74).
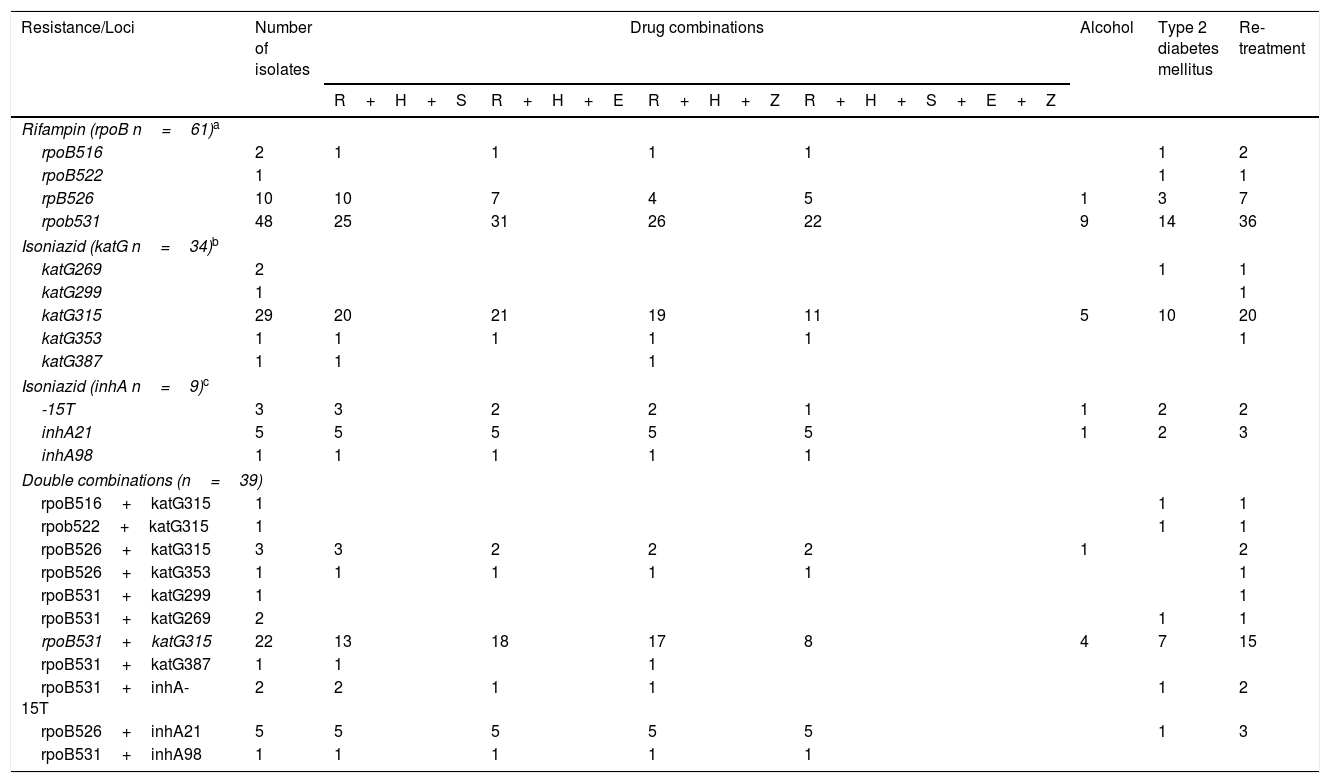
Table 4 shows the relationships between drug sensitivity profile, alcohol consumption, type 2 diabetes mellitus co-morbidity and type of treatment with single and double combination of mutations at katG/InhA and rpoB. Considering the 34 strains bearing a katG polymorphism, 29 (32%) had a mutation in katG315, of which eleven (32%) were resistant to all first line anti-tuberculosis drugs, 20 (58%) were in re-treatment and ten (29%) had type 2 diabetes mellitus. Of the 61 isolates with rpoB polymorphisms, 48 (80%) had a simultaneous mutation in rpoB531, of which 22 (36%) were resistant to all first line drugs, 36 (60%) were undergoing re-treatment and 14 (22%) had type 2 diabetes mellitus.
Combination of mutations and drug resistance profiles socio-demographic characteristics of MDR-TB isolates from Mexico.
| Resistance/Loci | Number of isolates | Drug combinations | Alcohol | Type 2 diabetes mellitus | Re-treatment | |||
|---|---|---|---|---|---|---|---|---|
| R+H+S | R+H+E | R+H+Z | R+H+S+E+Z | |||||
| Rifampin (rpoB n=61)a | ||||||||
| rpoB516 | 2 | 1 | 1 | 1 | 1 | 1 | 2 | |
| rpoB522 | 1 | 1 | 1 | |||||
| rpB526 | 10 | 10 | 7 | 4 | 5 | 1 | 3 | 7 |
| rpob531 | 48 | 25 | 31 | 26 | 22 | 9 | 14 | 36 |
| Isoniazid (katG n=34)b | ||||||||
| katG269 | 2 | 1 | 1 | |||||
| katG299 | 1 | 1 | ||||||
| katG315 | 29 | 20 | 21 | 19 | 11 | 5 | 10 | 20 |
| katG353 | 1 | 1 | 1 | 1 | 1 | 1 | ||
| katG387 | 1 | 1 | 1 | |||||
| Isoniazid (inhA n=9)c | ||||||||
| -15T | 3 | 3 | 2 | 2 | 1 | 1 | 2 | 2 |
| inhA21 | 5 | 5 | 5 | 5 | 5 | 1 | 2 | 3 |
| inhA98 | 1 | 1 | 1 | 1 | 1 | |||
| Double combinations (n=39) | ||||||||
| rpoB516+katG315 | 1 | 1 | 1 | |||||
| rpob522+katG315 | 1 | 1 | 1 | |||||
| rpoB526+katG315 | 3 | 3 | 2 | 2 | 2 | 1 | 2 | |
| rpoB526+katG353 | 1 | 1 | 1 | 1 | 1 | 1 | ||
| rpoB531+katG299 | 1 | 1 | ||||||
| rpoB531+katG269 | 2 | 1 | 1 | |||||
| rpoB531+katG315 | 22 | 13 | 18 | 17 | 8 | 4 | 7 | 15 |
| rpoB531+katG387 | 1 | 1 | 1 | |||||
| rpoB531+inhA-15T | 2 | 2 | 1 | 1 | 1 | 2 | ||
| rpoB526+inhA21 | 5 | 5 | 5 | 5 | 5 | 1 | 3 | |
| rpoB531+inhA98 | 1 | 1 | 1 | 1 | 1 | |||
Double polymorphisms were observed in 61 isolates. The most common combination was rpoBS531L (163 C/T)+katGS315T (276 G/C), which was observed in 22 strains. Seventeen (77%) individuals bearing this mutation also presented resistance to R+H+E, eighteen (80%) to R+H+P and eight (36%) were resistant to all first line anti-tuberculosis drugs. Fifteen individuals with this combination of mutations were also undergoing re-treatment, and seven (31%) presented type 2 diabetes mellitus.
DiscussionAmong the clinical variables observed in the population, it was found a high number of individuals older than 40 years old, this could have important implications in the transmission of TB, since it is an economically active population, and many of these individuals have households, whose members are at high risk to be infected by a DR- or MDR-Tuberculosis. The association with type 2 diabetes mellitus found was 35%, and keeps relation with national and state reports,18,19 this deserves special consideration because this association has been referred to as an important risk factor for the development of drug and multidrug-resistance,20 including population from this geographical location.21,22 This co-morbidity will undoubtedly need to be considered further in detail by health authorities.
Of the 74 individuals included in this study, 25% (19/74) were identified as new cases, signifying that they acquired TB infection with a strain that carries a previously developed resistance to rifampicin and isoniazid. Of the individuals analyzed, 34% (25/74), had resistance against all first line drugs, for this reason there are some concern that some of these isolates could also bear resistance against second-line drugs.
This fact evidences the magnitude and dispersion of these aggravated forms of TB in the region and illustrates the difficulties for adequate attention and effective control of MDR-TB. These data also demonstrate the urgent need to reinforce the systems of surveillance for drug resistant tuberculosis.
In the katG gene, six polymorphisms were observed in 46% (34/74) of the strains analyzed, the most frequent of which was katGG315A/C (39%). Previous descriptions of such change have been provided in Mexico,11,12,23 and in other countries such as Canada (46%)24 and Bangladesh (62%).25 However, important differences are observed when comparing these incidences with the 98% occurrence of this mutation in Kazakhstan,9 91% in Russia26 or 82% in Nepal.27 Of the remaining four polymorphisms observed in katG, three have not been previously reported (G137A, T398C, G491A) and therefore their possible participation in the process of resistance to isoniazid needs to be further explored.
It was not possible to carry out the sequence analysis of inhA promoter, so doubts about the polymorphisms that could be found remains, perhaps changes in this region could increase the low number of mutations related to resistance to isoniazid. However, the presence of several polymorphisms first described in the inhA region analyzed, such as inhAC-15T in the non-coding region, inhAA61G and inhAG294T(6%), was noteworthy.
Absence of any mutation in the katG and inhA genes was observed in 58% of the isolates. This is one of the highest frequencies described to date in isoniazid-resistant isolates from Mexico, and raises important questions about the participation of other genes such as inhA promotor, kasA, ahpC and oxyR3,7 that may influence the development of resistance to this drug in these isolates. Further studies are undoubtedly required to identify which genes and mutations might be involved in resistance to isoniazid in the Mexican isolates.
At least one mutation was found in the sequenced rpoB region in 82% of the isolates with resistance to rifampicin. This value was low in relation with other reports from Bangladesh, Nepal and Canada were the >95% of isolates had a mutation in rpoB,24,25,27 however this number is similar to those reported previously in Mexico11–13,23 and in other countries, such as China28 and Kazakhstan,29 partially confirming information related with the variations according to geographical areas. Of the eleven polymorphisms distinguished, the most frequent were rpoBS531L/W in 64% and rpoBH526D/Y/C/R/P in 13% of isolates. These coincide with previous reports from Mexico (53%)12,13,30 and other countries, such as Nepal (57.8%),27 Morocco (59%),31 Brazil (56%)32 and Bangladesh (52%).25 Nevertheless, 18% (13/74) of isolates did not show any change in the sequenced region further studies are required in order to identify polymorphisms outside the RDRR of rpoB or in other genes, such as rpoA and rpoC,33,34 that could help to explain the resistance to rifampicin present in such isolates.
Of the 74 isolates with confirmed phenotypic MDR-resistance, 18% (13/74) and 58% (43/74) presented no mutations in the rpoB and katG/inhA genes, respectively. This is an example of the discrepancies that exist between phenotypic and genetic methods (nucleotide sequencing) for molecular diagnosis of TB resistance and raises questions regarding the usefulness, not only of sequencing small fragments of genes and their potential use in diagnostic procedure, but also of other molecular tests based on identifying polymorphisms within specific genes, such as GeneXpert MTB/RIF, InnoLipaRif, MTBDRsl, etc. There is an indisputable need to develop more studies, in DR- and MDR-TB isolates from Mexico, relating to the identification of the polymorphisms that cause drug resistance against first and second line drugs and assessing the diagnostic value of the standard molecular diagnostic test.
One of the most important limitations of our study was the impossibility to determine the genotype characteristic of the recovered isolates. Nevertheless, it was possible to analyze the combination of changes and resistances; the data showed that 39 isolates (53%) featured eleven double combinations of polymorphisms at rpob, katG and inhA and important variations in the phenotypic resistance patterns (Table 4). A further limitation was related to the inability to follow those individuals carrying a strain with unusual mutations or with absence of polymorphisms in the genes studied, and to identify whether the infection was resolved or aggravated forms of resistance, such as pre-XDR or XDR, occurred.
This indirectly establishes that, more than the presence of a single clone or small groups of clones circulating in the area, there is an important diversity of drug-resistant isolates generating and circulating in the region, confirming previous descriptions made with isolates from this region.35 There is a recognized need to conduct genotypic characterization studies in order to evaluate in detail the diversity of genotypes that could be involved in the TB resistance phenomenon in the region. Considering the above, the genotypic characteristics of a group of MDR-TB isolates circulating in this region have recently been described,36 confirming the presence of two scenarios; isolates generated mainly by a mismanagement of a specific case and the presence of clones linked epidemiologically that actively participate in the dispersion of drug-resistant isolates. Undoubtedly, this information will be of great help for the management and control of these aggravated forms of tuberculosis by the health instances.
In conclusion, this study provides valuable information about mutations at rpoB, katG and inhA genes from clinical isolates of MDR-TB, in a region with an important prevalence of drug resistant tuberculosis in Mexico. These data further our knowledge of the molecular mechanisms related to drug resistance, and evidence the significant number of isolates that lack any mutation to explain their multidrug resistant character, supporting studies that show variations in the occurrence of mutations with relation with the geographical distribution of the MDR-TB isolates.4,6,7,10 Further studies need to be conducted in order of confirm the real participation of the new set of mutations found, especially in relation to isoniazid resistance. In these sense, the presence of new and unusual mutations, confirm previous observations related with the occurrence of variations in the polymorphisms associated with drug resistance in response to particular geographical, environmental and clinical conditions in which the drug resistant tuberculosis strains are emerging. This aspect need to be considered if new molecular diagnostic test want to be implemented in specific locations.
Undoubtedly, whole genome sequencing will be invaluable in terms of analyzing all of the genes that may be involved in developing drug resistance, including those related to second line drugs.33,37 This will have important implications for the diagnosis, treatment and monitoring of individuals recently diagnosed with TB, and suspicions of infection by with a drug-, multidrug- or extreme-drug-resistant isolate. All of this information will complete the panorama that allows the design and use of molecular tools for diagnosis of drug resistance with increased sensitivity and specificity for isolates circulating in specific geographical region.
Ethical approvalNo physical interventions occurred with the patients, and all information collected was confidential. Ethical issues derived from this study were overseen by the respective committee of the Public Health Institute of the University of Veracruz.
FundingThis research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Conflict of interestsNone declared.
None.