Serratia marcescens is a Gram-negative bacterium that is found in hospital environments and commonly associated with outbreaks in neonatal units. One S. marcescens isolate was detected from a bloodstream culture from a neonate in our hospital that was followed by an outbreak. The aim of this study was to describe the molecular epidemiology of a S. marcescens outbreak in the neonatal unit.
MethodsIn order to investigate the outbreak, weekly surveillance rectal swabs were submitted for culture from all patients admitted in this unit from August to September 2018. Environmental samples were obtained from potential sources in September 2018. Typing of isolates was performed by pulsed field gel electrophoresis (PFGE). In addition, we studied the in vitro activity of chlorhexidine against S. marcescens.
ResultsDuring this period, 146 infants were hospitalised in our neonatal unit, of which 16 patients had a S. marcescens-positive sample. A total of 36 environmental surveillance samples were collected, and one sample from a stethoscope from an incubator of a colonized baby was positive for S. marcescens. All the 18 isolates, including the isolate from the stethoscope, belonged to a single PFGE cluster. We found that very low concentrations of chlorhexidine, even with application times close to 0 achieved significant reductions in the amount of S. marcescens.
ConclusionA unique clone of S. marcescens caused this outbreak, including isolates from patients and from one stethoscope. The outbreak was controlled with the early implementation of specific control measures.
Serratia marcescens(S. marcescens) es una bacteria gramnegativa que se encuentra en ambientes hospitalarios y comúnmente aparece asociada a brotes en unidades neonatales. En agosto de 2018 se detectó un aislado de esta bacteria a partir de un hemocultivo de un paciente neonatal en nuestro hospital. El objetivo de este estudio fue describir la epidemiología molecular del brote de S. marcescens en la unidad neonatal.
MétodosCon el fin de investigar el brote, se enviaron semanalmente para cultivo muestras rectales de todos los pacientes ingresados en estas unidades de agosto a septiembre de 2018. Asimismo, se obtuvieron muestras ambientales de potenciales orígenes del brote en septiembre de 2018. Los aislados se genotipificaron mediante electroforesis de campo pulsado (PFGE).
ResultadosDurante este período, 146 lactantes fueron hospitalizados en nuestra unidad neonatal, de los cuales 16 pacientes tenían una muestra positiva para S. marcescens. Se recogieron un total de 36 muestras de vigilancia ambiental. Una muestra de un estetoscopio de una incubadora de un bebé colonizado resultó positiva para S. marcescens. Los 18 aislamientos, incluido el aislado de la muestra ambiental, pertenecían a un solo patrón de PFGE. Se realizaron experimentos in vitro con clorhexidina y se encontró que concentraciones muy bajas incluso con tiempos de aplicación cercanos a 0 lograban reducciones significativas en la cantidad de S. marcescens.
ConclusiónEstos datos confirmaron un único clon de S. marcescens en la unidad neonatal con aislamientos tanto de pacientes como ambientales. La implementación temprana de medidas de control específicas fue eficaz para limitar la transmisión nosocomial.
Serratia marcescens is an ubiquitous Gram-negative bacteria able to survive under extreme conditions.1,2
S. marcescens is a recognized cause of outbreaks in intensive care units,3 especially in neonatal intensive care units (NICUs)1,2,4,5 with high morbidity and mortality.6 During the last decade, several articles have been published describing outbreaks caused by S. marcescens identifying low birth weight and prematurity, prolonged hospital stay, invasive procedures, antibiotic use and mother's infections prior to delivery as risk factors for colonization and invasive infections.3,4,7
Several sources have been identified as reservoirs in hospitals, including the hands of medical staff, colonised infants and contaminated medical products and medical equipment, sinks, soap, and disruption of infection control measures, whereas no point source was identified in others.1,3,4,8 Therefore, the early detection of colonized or infected patients, or other sources of the outbreak and the prompt implementation of infection control measures are significant factors for better management of the spread of the organism.9,10
Typing of bacterial isolates can be particularly challenging and relies on the use of molecular biology tools such as pulsed field gel electrophoresis (PFGE), multilocus sequence typing, and whole genome sequencing.1 PFGE is the more commonly used typing technique and was applied to the investigations outbreaks.6
A S. marcescens isolate was detected from a bloodstream culture from a neonatal patient in our hospital in August 2018 that was followed by an outbreak. We describe the molecular epidemiology of a S. marcescens outbreak in the neonatal unit (NU), that included the NICU and the intermediate care unit, explain the control measures adopted, and evaluate the in vitro activity of chlorhexidine against S. marcescens.
Materials and methodsSetting and microbiological methodsIn our hospital the surveillance of the infection in the NU is carried out by a multidisciplinary team including paediatricians, the unit's nursing supervisor, microbiologists, and preventive medicine specialists. In addition, weekly team meetings are held and communication is constant.
In order to investigate the outbreak, surveillance rectal swabs were obtained twice a week from all patients admitted in the NU of the Miguel Servet University Hospital of Zaragoza (Spain) from August to September 2018 and culture results were reviewed. Informed consent was not required since all swabs were obtained for managing the outbreak, according to criteria for good clinical practice. Rectal swabs were cultured in MacConkey agar with cefotaxime (1μg/ml) and MacConkey agar with colistin (10μg/ml) as a specific culture medium culture Serratia spp. In addition, S. marcescens was isolated from clinical specimens using conventional bacteriological procedures.
Environmental samples were obtained in September 2018 from different rooms including sink drain-holes, tap water, tape, antiseptics (Hibiscrub®), medical equipment (stethoscope, ultrasound machine and contact gel, incubators, breast pumps, humidifiers), computer (keyboard and mouse), furniture surface, tables, sterile glove box, hands and cellular phones of healthcare workers (HCW), in an attempt to identify any possible source of infection.
Isolates of S. marcescens from clinical, screening, and environmental samples were identified by MALDI-TOF mass spectrometer (MALDI Microflex LT, Bruker Daltonics). Susceptibility testing of all S. marcescens was performed by MicroScan WalkAway (Beckman®) according to the European Committee on Antimicrobial Susceptibility Testing.11 The tested antimicrobial agents included were amoxicillin/clavulanic acid, ampicillin/sulbactam, aztreonam, cephalothin, cefepime, ceftazidime, ciprofloxacin, amikacin, gentamicin, imipenem, levofloxacin, meropenem, piperacillin, piperacillin/tazobactam, tetracycline, and trimethoprim/sulfamethoxazole.
Molecular epidemiologyPFGE was performed including one isolate from each patient. In patients with S. marcescens from screening and clinical isolates, both isolates were analyzed. PFGE was performed as previously described with some modifications.12 Briefly, DNA was digested with XbaI (Takara; Saint-Germain-en-Laye, France) and PFGE separation was performed on a CHEF-DR III System (Bio-Rad Laboratories, USA) at 14°C for 17h under the following conditions: initial switch time, 2.2s; final switch time, 63.8s; included angle, 120°; voltage gradient, 6V/cm. Salmonella serotype Braenderup H9812 was run in multiple positions in each gel to be used as the standard for the PFGE gels in accordance with PulseNet International protocols.13 DNA restriction patterns generated were interpreted according to the guideline.14 Cluster analysis was performed using the Dice coefficient with BioNumerics software version 6.0 (Applied Maths, Sint-Martens-Latem, Belgium). A coefficient of ≥0.80 was considered suggestive of possible clonal relatedness.
Study of the in vitro activity of chlorhexidine against S. marcescensWe studied the S. marcescens isolate obtained from the first infected patient detected in this outbreak. The inoculum was prepared in distilled water and adjusted to 0.6±0.03 on the McFarland scale. 2% aqueous chlorhexidine (CHX) (740-DES Miclorbic, Medichem S.A. Barcelona, Spain) was diluted in distilled water.
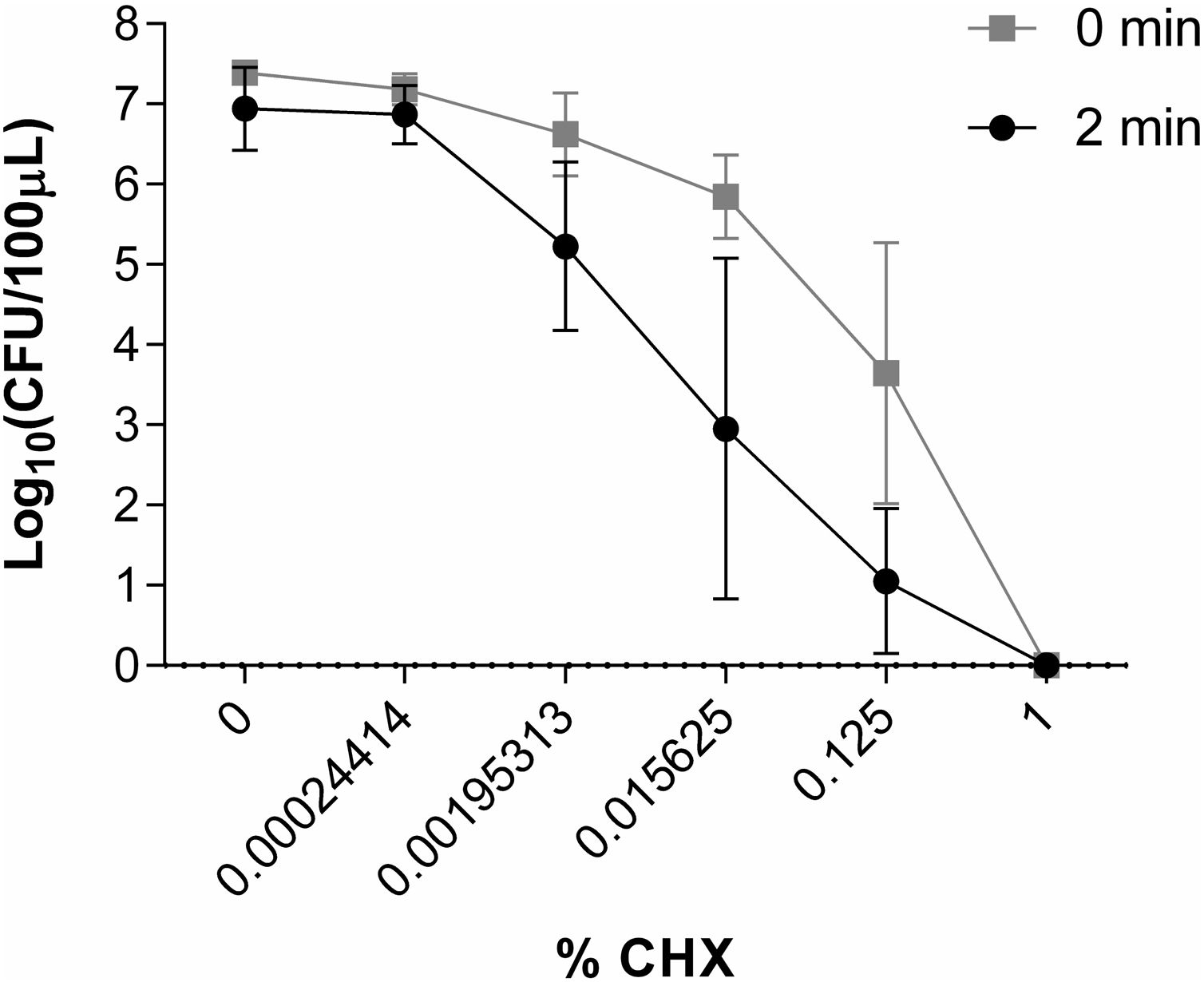
50μl of inoculum plus 50μl of CHX solutions were placed in different 1.5ml Eppendorf tubes. The concentrations of CHX studied were 1%, 0.125%, 0.015625%, 0.00195313% and 0.00024414% (corresponding to the dilution of CHX of 1/2, 1/16, 1/128, 1/1024 and 1/8192 respectively). Control tubes included 50μl of inoculum plus 50μl of water without CHX. Eppendorf tubes with the different CHX concentrations were prepared in duplicate: one was inoculated immediately, and another was incubated at 35°C for 2min before being inoculated. Samples and controls were cultured on blood agar and incubated overnight at 35°C. The activity of CHX was assessed by counting the number of colony-forming units per 100μl (CFU/100μl) using a Flash and Go automatic colony counter (IUL, S.A, Spain) and comparing the results of the samples with the controls. All experiments were carried out at least 3 times. A reduction in the number of CFU/100μl of 6 log10 was considered indicative of bactericidal activity.
ORION statementWe have verified that all the points of “The ORION statement” are fulfilled to ensure the quality and transparency of this outbreak report.15
Ethical approvalThe study was approved by the local ethics committee (Comité de Ética de la Investigación de la Comunidad de Aragón, reference number: 01/2019).
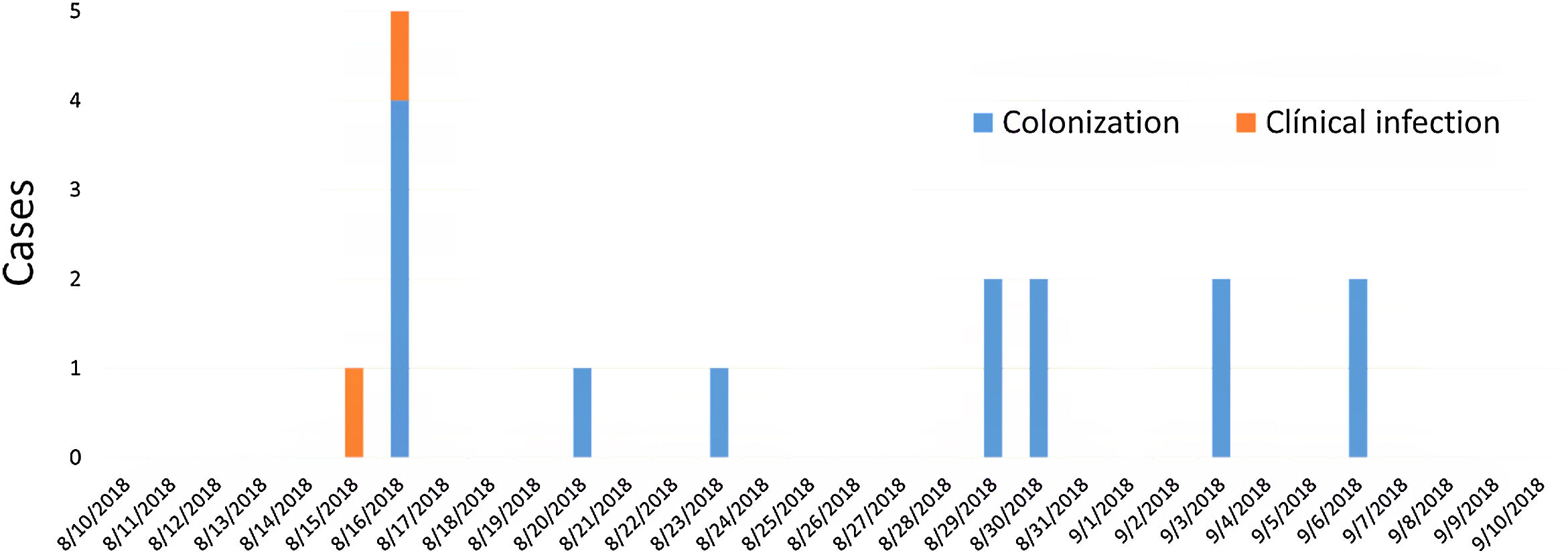
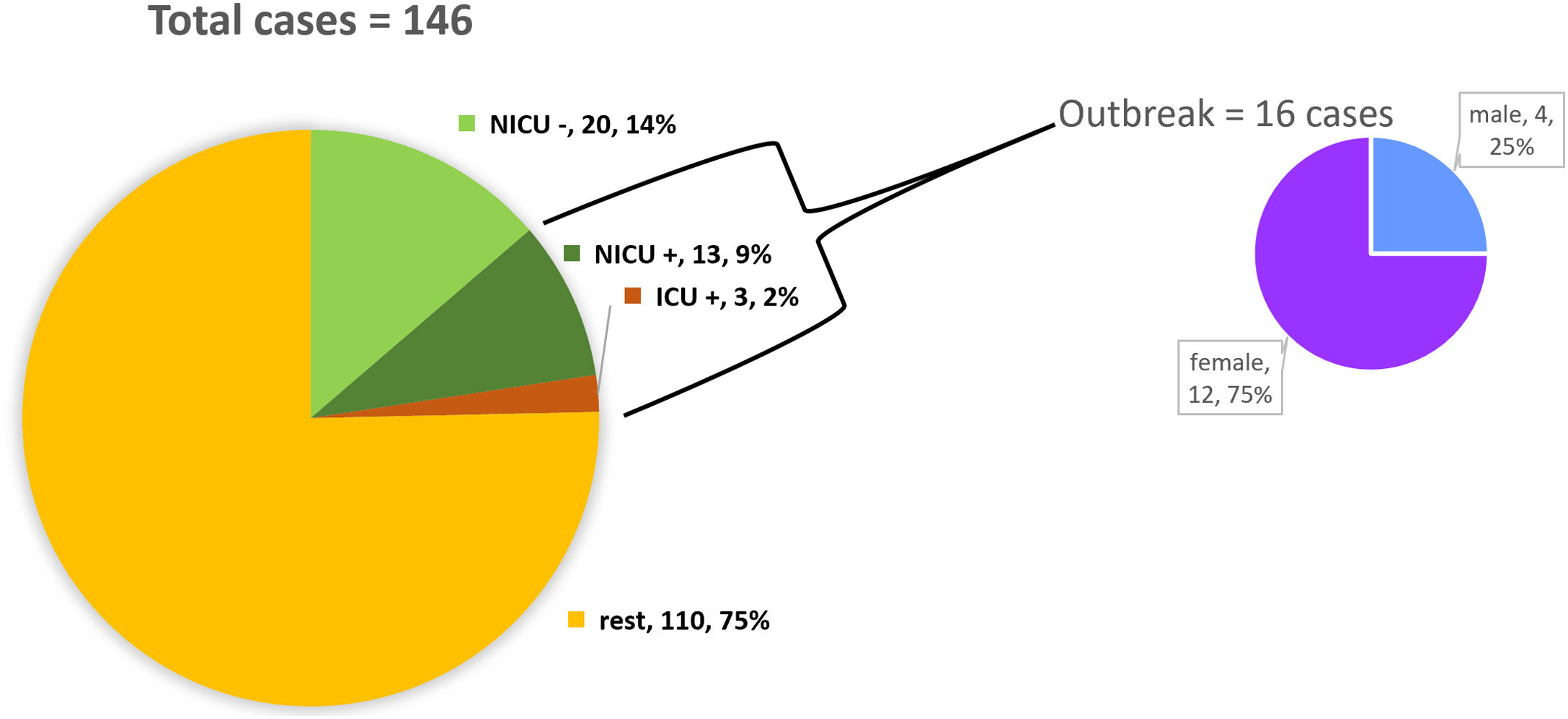
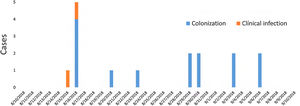
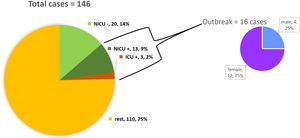
ResultsDescription of the outbreak and microbiological resultsFrom August 1 to September 30, 2018, a total of 146 infants were hospitalised in our NU. Among them, 33 were hospitalized in the NICU, and all of them were screened using rectal swabs. During this period, 16 (11%, 14 colonized+2 infected) patients colonized or infected with red pigmented S. marcescens were identified (Fig. 1). Thirteen of the patients were from NICU and three from Intermediate Care Unit, yielding an attack rate of 39.4% in the NICU. Out of 16 patients 4 (25%) were males and 12 (75%) females (Fig. 2). Fifteen isolates were obtained from screening samples and two from clinical samples. One patient had two isolates, one epidemiological and one clinical. The first two cases detected in August were two patients with bacteraemia obtained in two consecutive days, one of whom died. Both patients had low birth weight, prematurity, prolonged hospital stay, and invasive procedures.
A total of 36 environmental surveillance samples were collected, and S. marcescens was isolated from one stethoscope from an incubator of a colonized baby. None of the cultures obtained from hands of HCW yielded S. marcescens. However, samples from tap water, sink drain-holes, computer keyboard and mouse, hands and cellular phones of HCW, ultrasound machine revealed the presence of several microorganisms, including Staphylococcus spp., Pseudomonas spp., Pantoea spp., Klebsiella spp., Enterobacter spp. and Stenotrophomonas spp.
Antibiotic susceptibilityAll isolates showed an ampicillin/sulbactam, amoxycillin-clavulanic acid and cephalothin resistance phenotype, characteristic of the chromosomal inducible AmpC beta-lactamase characteristic of this species. In addition, isolates were colistin and tetracycline resistant. The isolates were susceptible to imipenem, meropenem, ciprofloxacin, levofloxacin, gentamicin and trimethoprim/sulfamethoxazole. None of the isolates were extended spectrum beta-lactamase(ESBL)- or carbapenemase-producer.
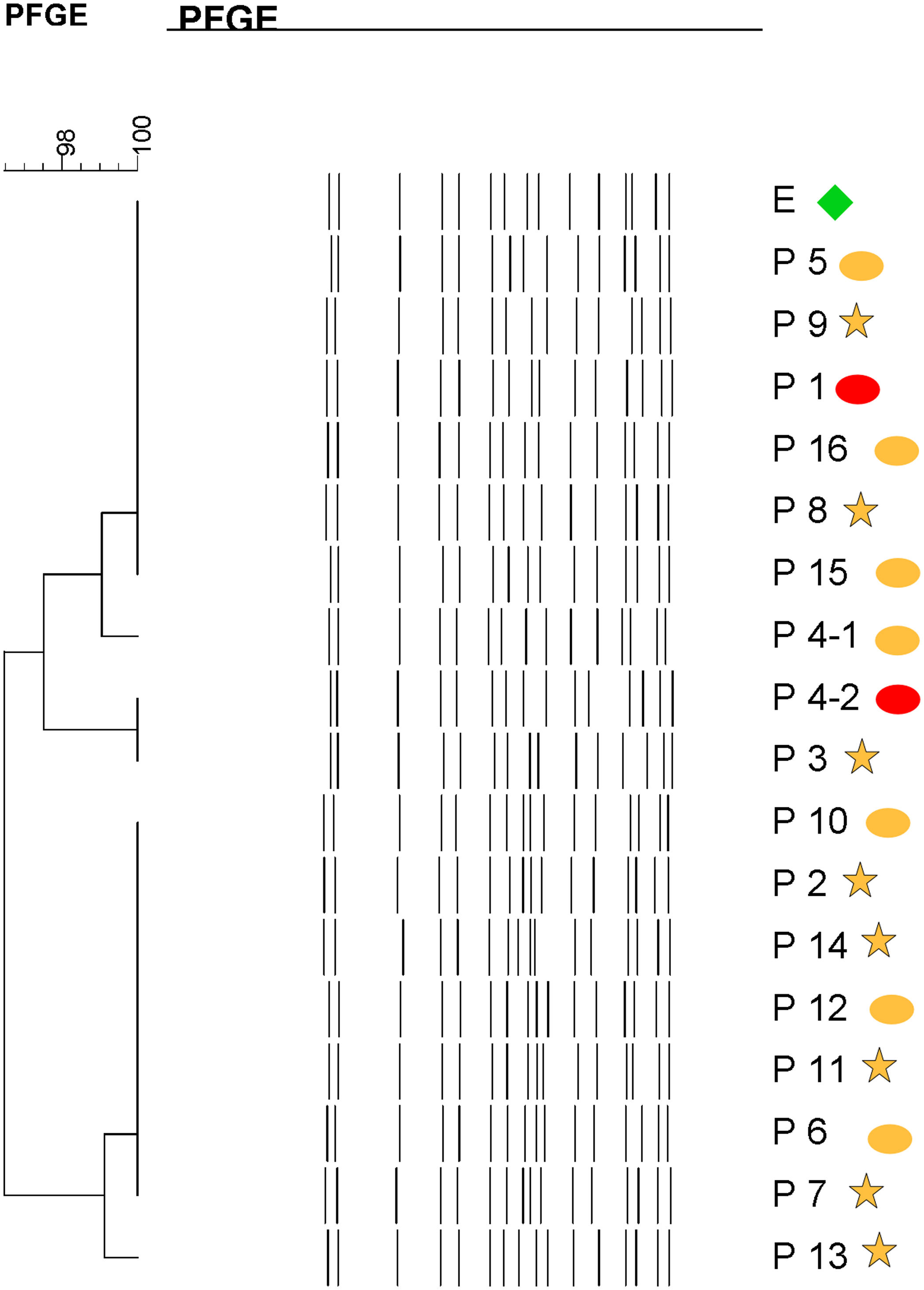
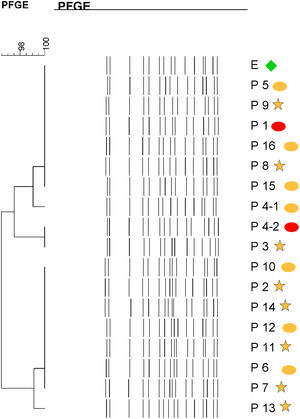
Molecular epidemiologyMolecular typing by PFGE was performed on 18 S. marcescens isolates, including one environmental isolate. All S. marcescens clinical, epidemiological and environmental isolates had>90% similarity, belonging to a unique PFGE cluster (Fig. 3). This clone was responsible for the outbreak from August to September.
Dendrogram showing results of PFGE profile of environmental, clinical and epidemiological isolates of S. marcescens in the neonatal unit. Environmental (E, green diamond symbol); patients (P); Ellipse symbol indicates that the sample comes from the neonatal intensive care unit. Star symbol indicates that the sample comes from the intermediate care unit. Red colour indicates blood sample and the orange colour indicates rectal sample.
The outbreak was controled by specific screening surveillance of neonates, reviewing all healthcare procedures, reinforcing standard infection control procedures, including contact precautions, cohorting infected/colonized neonates and nursing, and disinfecting environmental surfaces. Compliance with hand hygiene in all contacts with patients and their environment through training of personnel in the unit, observation of compliance and feedback from personnel was reinforced. We performed microbiological cultures from the hands of the personnel and results were presented.
Possible sources were disposed and renewed, including disinfectants, soaps and soap dispensers, cotton and tissues, and even temporary closure of the affected department in order to replace sink drains, to paint the entire unit and to replace damaged surfaces and furniture in the NICU.
The screening of neonates continued after the control of the outbreak. No other infections caused by S. marcescens were diagnosed, and the colonization rate decreased significantly after implementation of infection control measures. Nine cases of S. marcescens colonization were found in the NU, two of them in the NICU, during 6 months after the outbreak but, at that time, the outbreak was over, and PFGE analysis demonstrated that these neonatal isolates were genetically different. In addition, colonies of these 9 isolates did not produce a red pigment.
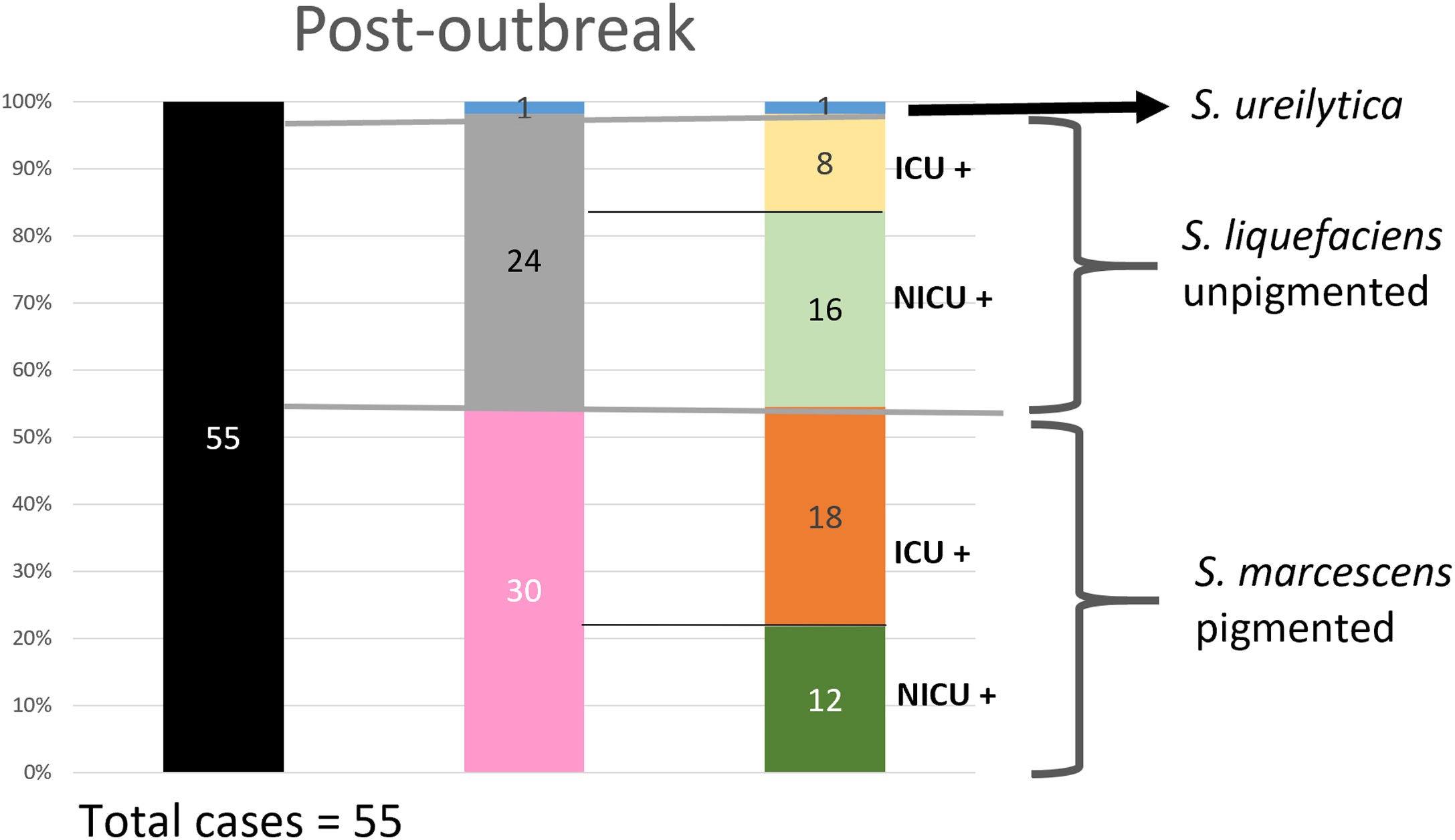
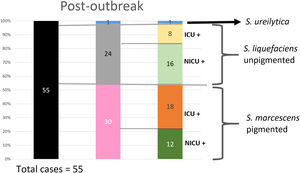
Analysis of the epidemiological situation in the neonatal unit after the outbreakDuring the 13 months after the outbreak, a total of 55 patients colonized by Serratia spp. were detected: 30 patients (12 in NICU and 18 in intermediate care unit) by non-pigmented S. marcescens; 24 patients (16 in NICU and 8 in intermediate care unit) by S. liquefaciens; and 1 (1 in NICU) by S. ureilytica. No cases of bacteraemia were detected during this period (Fig. 4).
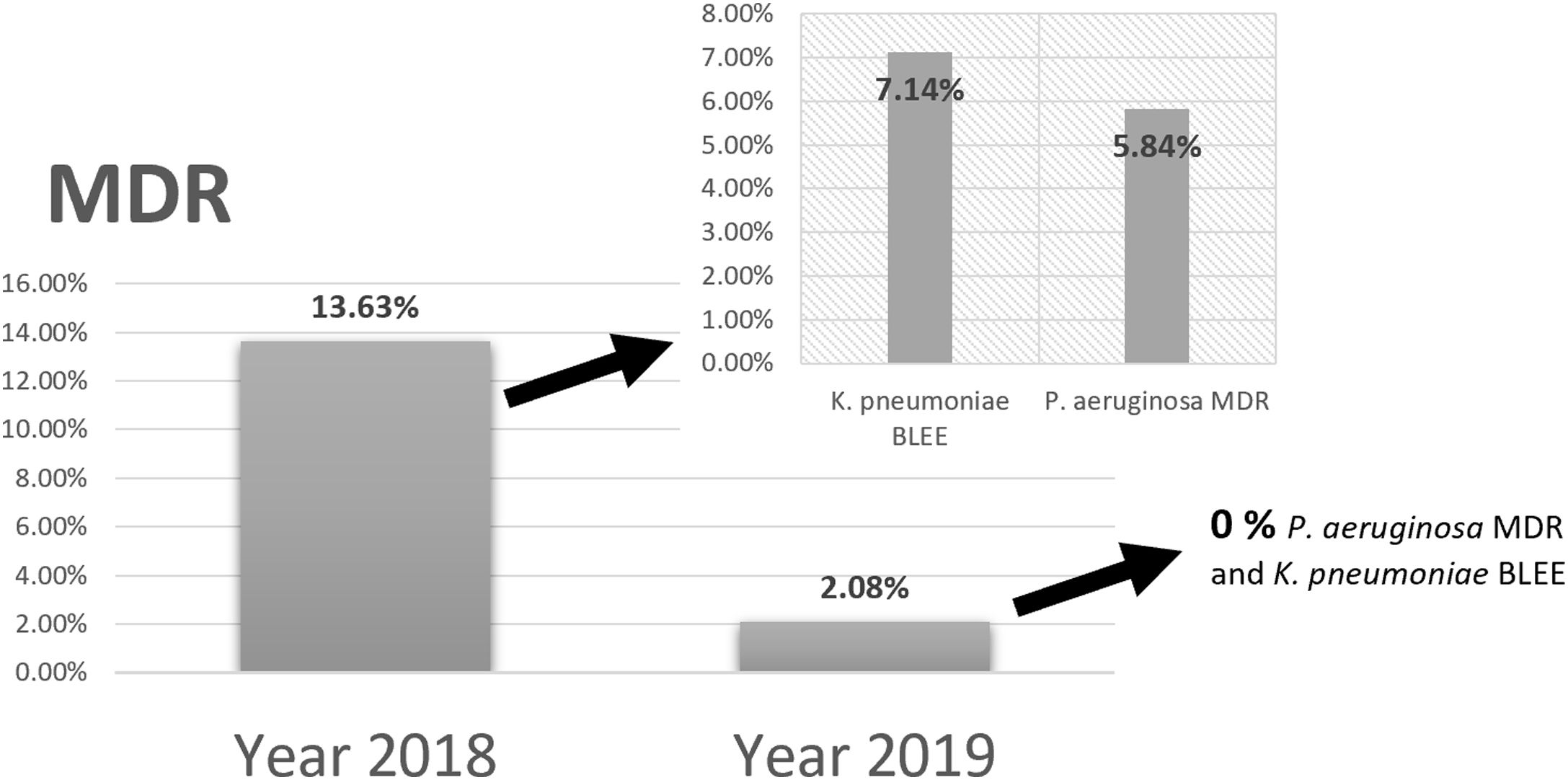
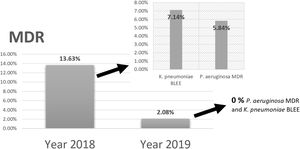
Comparing the results of epidemiological surveillance of 2018 with those of 2019 in NICU, there was a decrease from 13.63% to 2.08% in the percentage of patients colonized by multidrug resistant bacteria (MDR). The rates of 5.84% (P. aeruginosa MDR), and 7.14% (ESBL-K. pneumoniae) in 2018, decreased to 0% in 2019 (Fig. 5).
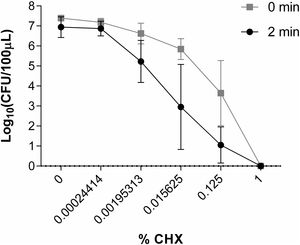
Study of the in vitro activity of chlorhexidine against S. marcescens1% CHX effectively inhibited S. marcescens, resulting in 6 log10 reduction in bacterial growth in all assays (Fig. 2). CHX at concentration of 0.00024414% was not effective when was applied both for 2min and for a time close to 0. Neither was the concentration of 0.00195313% nor 0.015625% applied for a time close to 0 to reduce the number of CFU/100μl. However, these concentrations of CHX when applied for 2minutes reduced the amount of CFU/100μl by ∼1 log10 and ∼3.5 log10, respectively. CHX at a concentration of 0.125% reduced the amount of CFU/100μl significantly when applied both for 2minutes (∼5.5 log10) and for a time close to 0 (∼3.5 log10) (Fig. 6).
DiscussionS. marcescens is a well-known causative agent of hospital infections involved in outbreaks at NICU that are characterized for its quick spread, causing severe complications.9,10S. marcescens outbreaks represent a serious challenge in NICU9 being the third most frequent pathogen involved in outbreaks in neonatal facilities.5 Here, we report a genetically confirmed monoclonal outbreak of S. marcescens involving 16 patients in the NU of the Miguel Servet University Hospital.
In neonates, the main risk factors for the acquisition of Serratia spp. infection are: prematurity and low birth weight, length of stay, exposure to invasive devices such as central or percutaneous venous catheter, and use of antibiotics.5,7 These risk factors were present in the two infants with bacteraemia in our outbreak.
Rates of mortality due to S. marcescens bacteraemia can vary significantly.5 In a report by Kim et al. none of the patients died from S. marcescens infection,16 Polilli et al. reported a mortality rate of 40%,17 and we observed 2 cases of bacteraemia with one fatal outcome, being the mortality rate 50%.
Specific surveillance cultures and molecular typing of isolates has proved to be a very useful tool for identification and evaluation of S. marcescens outbreaks in hospitals and, thus, implementing effective preventive measures when the sources are identified.3,10 Although whole genome sequencing of S. marcescens isolates have recently become available,5 PFGE is a more accessible useful method to identify outbreaks of S. marcescens.10 PFGE allowed us to demonstrate that all cases belonged to a single clone including the isolate obtained from the stethoscope. This is in contrast with other investigations where more than one clone has been identified during S. marcescens outbreak investigations.4,6,9
In outbreaks, extensive environmental microbiological investigations are needed to identify potential sources and reservoirs.17 Some studies have identified sources or reservoirs during S. marcescens NICU outbreaks.4 The reservoirs most frequently associated in these cases are: washbasins,18 tap water, air-conditioning systems,19 milk,6,10 soap-dispensers,17,20 baby shampoo,21 disinfectants,22 parenteral nutrition,8 prefilled syringes containing heparin or normal saline,23 multi-dose vials of heparinized-saline,24 extrinsic contamination of the parenteral narcotic fentanyl by a HCW,25 neonatal incubator,26 and hands of HCWs.27 In other outbreak studies, the reservoir and mode of transmission has remained unknown.7
The initial potential source and how the S. marcescens isolate was introduced in the NU remain unclear. It is possible that the index case was the first neonate with bacteraemia admitted on August in the NICU, and later, other neonates in the intermediate care unit were infected. S. marcescens was only isolated from one stethoscope and this isolate showed the same PFGE profile than isolates from patients. However, despite the association, we were unable to state definitively whether the stethoscope was the cause of the outbreak or had been contaminated as a result of it. Although S. marcescens was not found on the hands of staff, this could be the most likely way of spread. In our study, samples from tap water, sink drain-holes, computer keyboard and mouse, hands and cellular phones of HCW and ultrasound machine revealed the presence of several microorganisms suggesting noncompliance with hand hygiene by HCW. The presence in the NU of many neonates colonized at the gastrointestinal level could indicate that transient colonization from neonates may be more important than environmental contamination. Most of the published studies reported that colonized and infected infants are the most important reservoir of S. marcescens,3,9,10 and the contaminated hands of HCWs are probably the main vehicle of transmission of the pathogen.5,9
It is essential to remain vigilant while maintaining strictly control strategies. In this regard, the early diagnosis of patients who are colonized or infected and the rapid implementation of infection control measures are key factors in curbing the spread of S. marcescens.5,7 Since contaminated hands of HCW could be the most important vehicle of S. marcescens transmission when gloves are not used, training staff in proper and rigorous hand-washing and the use of gloves when assisting and caring for neonates was an important control measure to adopt.5 In our NU, the control measures, including cohort nursing, the temporary closure of the unit and the use of the ultraviolet radiation disinfection system used, may have contributed to the control of S. marcescens spread in the ward.5,6
The spreading of S. marcescens stopped after the implementation of control measures in the NU. There are several reports on secondary and even tertiary waves of transmission.6 Fortunately, our outbreak was limited to a single peak of infection or colonization, and there was no recurrence. Moreover, the temporal and definitive implantation of prevention and surveillance measures applied for S. marcescens in the NU seemed to have a preliminary positive impact on the cross-transmission with all MDR Gram-negative bacteria. According to the study of Casolari et al.,3 in our study, the implementation of infection control measures lowered the rates of ESBL-producing K. pneumoniae and MDR Pseudomonas aeruginosa.
CHX is effective against a broad-range of microorganisms, including MDR.28 Several studies have demonstrated its effectiveness as a skin disinfectant.29,30 On the other hand, some authors suggest that the spread of S. marcescens, can be facilitated by resistance to disinfectants. De Frutos et al. described an outbreak of S. marcescens infections linked to the use of 2% CHX.22 Dawczynski et al. reported disinfectant tolerance of S. marcescens.4 However, our results indicate its complete inactivation with 1% CHX even with application time is close to 0. Very low CHX concentrations (0.015625–0.125%) with only 2min of application achieve significant reductions around≥3 log10 in the number of CFU/100μl in the amount of S. marcescens in vitro. These results could support the effectiveness of use of 2% CHX against S. marcescens in order to reduce the spread. However, more experiments would be necessary to perform with low concentrations of CHX before its clinical application.
In conclusion, our results confirmed an outbreak caused by a unique clone of S. marcescens in the NU, including isolates from patients and one environment isolate, suggesting the importance of surveillance procedures for infection control and the importance of molecular biology in helping to understand spread of microorganisms. Rapid hospital implementation and adherence to infection control measures, and screening of patients seemed to be effective in overcoming this S. marcescens outbreak.
Contributors’ statementM.I. Millán-Lou participated in the conceptualization and design of the study, carried out the experiments and drafted the initial manuscript, C. López and J. Bueno were in charge of collecting samples, culturing and studying the sensitivity and participated in drafting the article, V. Pérez-Laguna, participated in the design of experiments with chlorhexidine and carry out the experiments, draf the article and was in charge of editing and sending it, C. Lapresta and M.E. Fuertes established and was in charge of the analysis of the control measures and reviewed the manuscript, S. Rite coordinated the unit's staff, promoted and facilitated the taking of control measures and revised the manuscript, M. Santiago and M. Romo collected samples, carried out the initial analyses and participated in revise the manuscript S. Samper provided equipment, assisted in data analysis and revised the manuscript, A. Cebollada performed the bioinformatic analysis of the data and participated in its interpretation, J. Oteo-Iglesias participated in the typing of the isolates and collaborated in the comparison of results from the two laboratories and reviewed the manuscript. A. Rezusta was in charge of conceptualizing and designing the study in mostly, organized the laboratory resources, obtained the resources for typing, participated in data analysis and in writing the manuscript. All authors approved the final manuscript as submitted and agree to be accountable for all aspects of the work.
Funding sourceMaría Isabel Millán-Lou received funding from the Instituto de Salud Carlos III (grant JR15/00011). The authors acknowledge financial support by IIS-Aragón (Instituto de Investigación Sanitaria Aragón), the Departamento de Ciencia, Tecnología and Universidad from the Gobierno de Aragón, Spain (Project DGA-European Social Fund (FSE)/Grupos consolidados, B10-17R. “Microbiología de las infecciones de difícil diagnóstico y tratamiento”) and FEDER (Fondo Europeo de Desarrollo Regional. Aragón 2014–2020: “Construyendo Europa desde Aragón”).
Conflict of interestThe authors have no conflicts of interest to disclose.