Introduction
In recent years, the contribution of chemotherapy to infection by the human immunodeficiency virus (HIV) has been extremely significant with regard to survival and improvement of quality of life. Nevertheless, a cure for this infection remains an uncertain objective as the virus cannot be completely eliminated, but remains latent and able to reactivate in a small cell reservoir1,2. Consequently, alternative therapies, known as gene therapy3, have been applied to this problem these include marrow transplant, cell therapy, immune stimulation, use of therapeutic vaccines and genetic interventions.
Given that HIV integrates into the host cell genome, aids can be considered as an acquired genetic illness4. Despite the fact that attempts have been made to genetically cure the provirus using excision techniques5, most anti-HIV gene therapy has tried to introduce an antiviral gene into the cell to prevent infection or inhibit viral expression. David Baltimore originally named this process "intracellular immunization"6 and it is the ultimate objective of gene therapy. This discipline is the fruit of extraordinary advances in molecular pathology and genetics, and generated many expectations from its birth to the 1990s, although they were not always accompanied by convincing clinical results7. The real situation is similar to that of HIV infection, in that the development of gene therapy is much more complex than expected. The task of introducing and expressing genes in somatic cells requires a detailed knowledge of the molecular bases of the disease and an improvement in gene transfer techniques8. As we can see below, these advances mean that gene therapy has seen moments of intense euphoria and disappointment, and the development of formidable molecular techniques, such as the RNA interference mechanism.
Anti-HIV molecular strategies
HIV is a complex retrovirus which, apart from the products of the genes gag, pol and env of simple retroviruses, uses six accessory proteins whose function is essential for the replication of the virus and completion of the infectious cycle. Some of these products, mainly Tat and Rev, together with the receptors necessary for cell infection, have been the main targets of molecular interventions.
Tat protein
Tat is a potent transactivator of the LTR promoter of HIV-1 and is essential for the replication of the virus9. Two mechanisms have been used to interfere with Tat: in the first, an anti-sense RNA corresponding to the TAR region is expressed in such a way that its binding prevents integration of Tat into this structure and therefore its function4. Another blocking strategy of Tat involves the expression of large quantities of RNA fragments corresponding to TAR, by using them as a decoy to reduce the number of effective Tat molecules which can bind to retroviral TAR10.
Rev
The carboxy-terminal portion of Rev contains a region rich in leucin residues which works as a nuclear exportation signal via interaction with cellular transport machinery11. Different Rev transdominants have been constructed, but the most widely used is known as RevM10, which contains a mutation precisely in the region of interaction with cell proteins12. RevM10 has been used by several groups in human clinical trials13,14 with very limited results15,16. RevM10 has some very attractive characteristics from the point of view of its use as an antiretroviral: it has no cell toxicity, it is very immunogenic, and the development of resistant variants seems more difficult than when we use an enzymatic target (although they have recently been induced in vitro17).
Receptors
Isolates used during the initial phases of infection invariably use the CCR5 molecule as a co-receptor18. The significance of this fact increased when it was discovered that some subjects who were homozygotic for a 32-bp deletion in the CCR5 gene19,20, present in 1% of Caucasians21, were resistant to HIV-1 infection. Moreover, patients carrying this deletion heterozygotically, even though they could become infected, had a slower clinical outcome22,23. It seems clear that interference with the expression of CCR5 and CXCR4 could have important therapeutic benefits. Nevertheless, even though complete destruction of CCR5 (which is the case of subjects who are homozygotic for D32CCR5) does not seem to be associated with any immunodeficiency, CXCR4 blockage could be more problematic since, at least in mice, destruction of CXCR4 has severe consequences on hematopoiesis and cerebral development, although its destruction in post-developmental stages could be better tolerated.
An alternative for blocking co-receptor expression in the cell membrane is to use of the membrane's own natural ligands modified in such a way that they can bind intracellularly to their receptors and at the same time be blocked in the endoplasmic reticulum (ER), thus limiting accessibility of the virus for this co-receptor. These molecules which can inhibit transport of its receptors to the cell membrane are known as "intrakines"24. The intrakines whose antiviral potential is being evaluated are based on RANTES, MIP-1α and MIP-1β for CCR5 (MIP-1β is completely specific for CCR5 whereas RANTES and MIP-1α can also block other receptors) and SDF-1 for CXCR5.
As well as the intrakines, other strategies can help us block expression of these co-receptors at a different level, for example by using ribozymes. Ribozymes catalyze the specific cut of RNA sequences and, due to their efficiency and the simplicity of their design, have become interesting tools for the selective suppression of gene expression25. Recently26, the design and function of the "hammerhead" ribozyme, RzR5-76, has been studied. This ribozyme specifically hydrolyzes the mRNA of CCR5 and the results of an in vitro transfection model indicate that its catalytic activity makes possible an important reduction in the expression of the co-receptor CCR5 on the cell surface26. Similarly various strategies have been used against the CD4 receptor, including the manufacture of CD4 chimeras with retention signals in the ER27.
Tools for genetic transfer
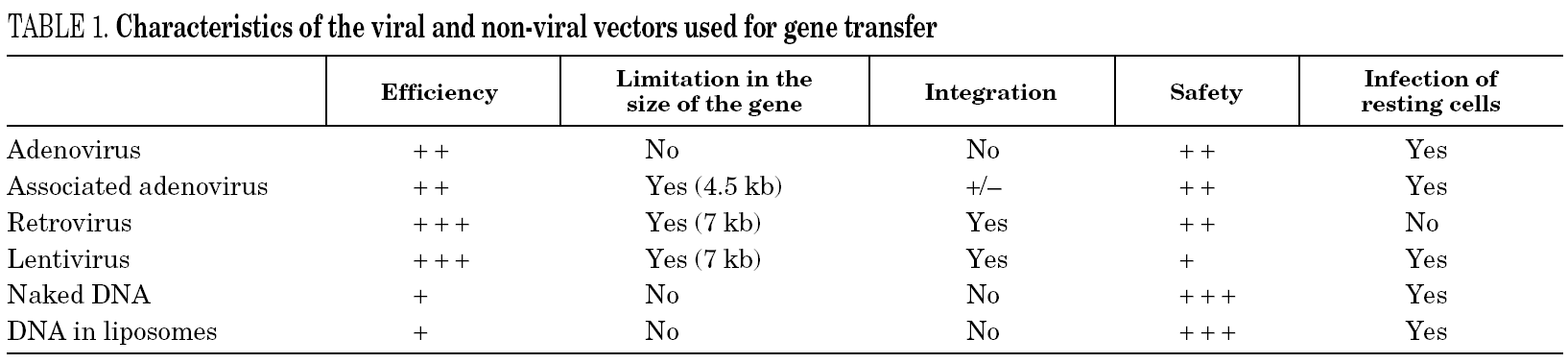
As a realistic therapeutic alternative, gene therapy is limited by the development of genetic vehicles, or effective and secure vectors, with which we could bring about the restoration of a defective gene or, as is the case with aids, transfer antiviral resistance to susceptible cell populations. There are different ways of introducing DNA into a cell (table 1), but the most widely used at present involves vectors based on recombinant viruses, especially retroviruses.
Retroviruses
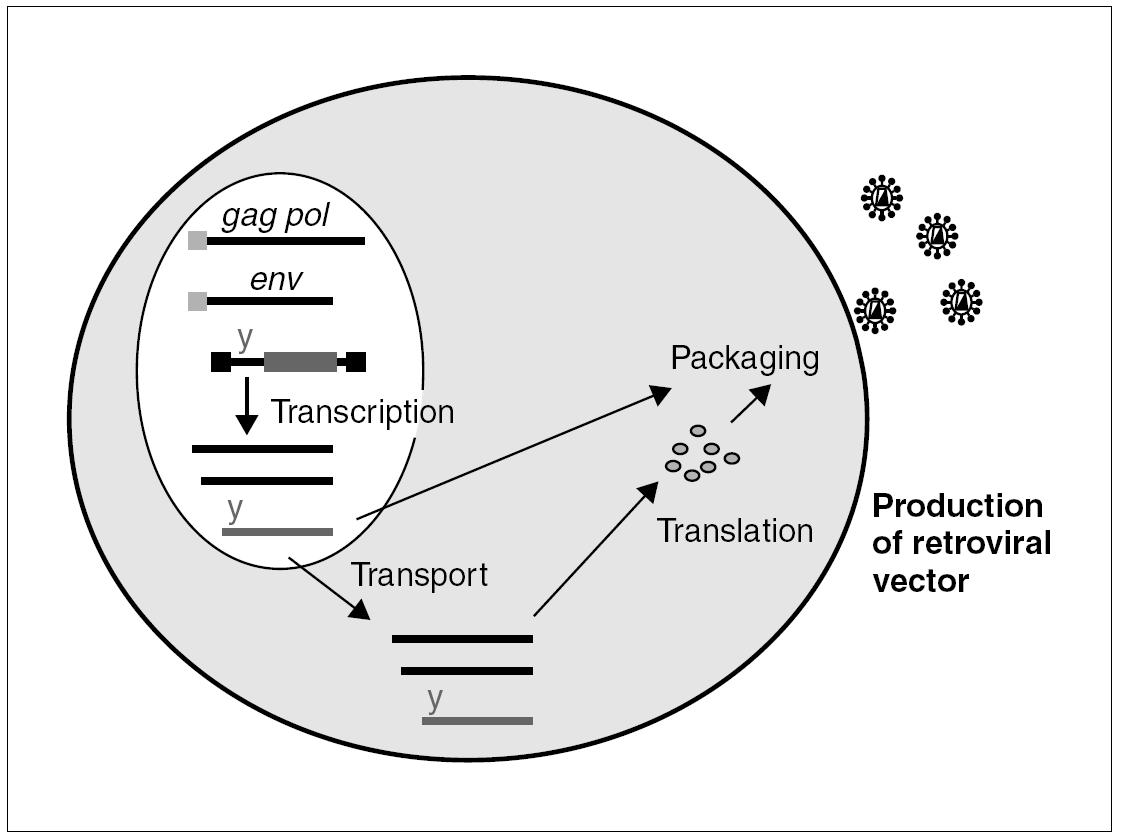
The properties which make retroviruses so dangerous from a pathogenic viewpoint are precisely those which make them very attractive as vectors in gene therapy: they infect cells efficaciously and, once integrated in the cell genome, especially if the cell has no structural proteins, their expression is immunologically silent. A recombinant virus is manufactured by using the simple murine oncoretrovirus, the Murine Leukemia Virus (MLV) (fig. 1), to eliminate most of the gag, pol and env sequences. These sequences encode their structural products and are substituted by the sequence of the gene we wish to express. The remaining structural components of the viral particle are contributed in trans, directed by a heterologous promoter. In this way, we can produce defective retroviral particles with the capacity to infect cells and integrate into their genome, albeit without the capacity to produce a new infective cycle (given that the genome lacks structural genes).
Figure 1. Packaging cell for the production of recombinant retroviruses: the structural components, gag, pol and env, of the retroviral particle and the genomic RNA penetrate the cell by transfection of different plasmids. All of these components are packaged and generate a defective infectious particle, with no structural genes, which will integrate the transgene into the target cell.
Lentiviral vectors
Despite our experience with vectors based on the murine oncoretrovirus, their use in therapy is limited by their inability to transduce cells which are not actively dividing, such as muscle, nerve or hematopoietic cells.
This is the main reason for using vectors based on lentiviruses which, thanks to the karyophilic properties of their pre-integration complexes, manage to efficaciously infect resting cells28,29. The first lentiviral vectors were derived from HIV-1 itself28,29 and other lentiviruses: HIV-230, simian immunodeficiency virus (SIV)31 and felin immunodeficiency virus (FIV)32 have since been used. Obviously, vectors based on pathogenic agents must be used under conditions of extreme biosafety, which makes the recombination and generation of competent variants impossible33-35. For example, the design of lentiviral vectors currently includes a minimum of original sequences, barely 40%, and introduces safety mechanisms such as auto-activation of the promoter (LTR)33,36,37.
One of the most attractive applications of lentiviral vectors is transduction of precursors, which are mother cells with a huge capacity for differentiation and auto-renovation. In this way, we can guarantee expression of the transgene in differentiated cells, for example HIV-resistant CD4+ lymphocytes in the case of aids, which would originate unbounded from this population of mother cells. Animal study models of hematopoietic reconstitution in NOD/Scid, a mouse with severe combined immunodeficiency and natural Killer (NK) activity deficit38, are very promising. It has been shown that repopulation of the whole human hematopoietic system from umbilical cord blood precursors which have been transduced ex vivo with a genetic marker by means of a lentiviral marker, and that expression of the marker is maintained for months39. Similarly, maintained expression and transduction have been demonstrated with quiescent cells in other tissues, such as brain28,29 and retina40 in experimental models using lentiviral vectors.
Nevertheless, vectors based on HIV-1 must be used with caution, and notwithstanding the results obtained in pre-clinical studies, the demands of biosafety logically delay their application in humans. Foreseeably, HIV-1-infected patients will be the first to use lentiviral vectors. Even though the defective retroviral vector expressing the resistance marker can in fact be rescued by the natural virus, this could have the added benefit of expanding the resistance gene33.
Situation of gene therapy studies in HIV infection up to the year 2000
As far as the year 2000, a considerable number of clinical studies using gene therapy for the treatment of HIV-1 infection were authorized. These phase I and II studies aimed to confirm the safety of the procedures and the absence of toxicity in genetic interventions on human cells. The clinical efficacy results using these first-generation retroviral vectors have been very limited. The most significant study to date with published data16 has enabled us to show, in HIV-1-infected patients, a more prolonged survival of CD4 lymphocytes which express the antiviral gene RevM10 in comparison with those which express a non-functional control. However, this selective advantage of the protected cell population did not achieve a significant reduction in viral load, probably due to the fact that the quantity of transduced CD4 cells (around 0.1% of the circulating cells) is too small to modulate expression of the illness.
Problems in the use of retroviral vectors
In April 2000 the Alain Fischer and Marina Cavazzana-Calvo group of the Hospital Necker in Paris reported the first gene therapy study with unequivocal therapeutic success. The clinical protocol was carried out on a group of 10 children with a form of Severe Combined Immunodeficiency linked to chromosome X (SCIDX1) due to the absence of the subunit g of the receptors of IL-2, 4, 7, 9, 15 and 21. Therapy involved the repositioning of the deficient receptor (IL2Rgc) in autologous CD34 hematopoietic cells using a retroviral vector. After re-infusion of the modified cells, 9 of the 10 children presented a rapid functional and quantitative repopulation41. Cure by gene therapy had been achieved. Nevertheless, the extraordinary optimism generated by these results turned to disappointment when it was shown that two of the children were developing leukemia42. The study of the leukemic cell population confirmed that, in both cases, the retrovirus which expressed the IL2Rgc gene had integrated close to the promoter of the lymphocyte oncogene, LMO243. These severe secondary effects led to a moratorium in testing with retroviruses in most countries. In the case of IL2Rg deficiency, there may have been a sequence of unfortunate circumstances: over-expression of IL2Rg may itself be oncogenic and, given the high number of cells modified, increases the probability of insertion near another oncogen. It is precisely these cells which have a greater potential for selection during repopulation and which can eventually develop leukemia44.
Integration of retrovirus and HIV into the human genome was considered random. However, at present, we know that it occurs in clearly preferential zones (hotspots). The availability of the human genome sequence, together with a greater capacity to analyze insertion regions thanks to automated sequencing and amplification, have enabled us to determine that HIV prefers to enter active genes45. In the case of the murine retrovirus, the preferential zones appear to be right at the proximity of promoters46, which means that greater caution must be exercised when using these vectors47.
RNA Interference
The recent description of the mechanism known as RNA interference (RNAi) has aroused enormous interest and has quickly become one of the most active areas of biological research. This phenomenon involves specific silencing of the expression of certain genes by short fragments of double-stranded DNA (dsDNA). In 1998, Fire et al, using the nematode Caenorhabditis elegans as an experimental model, confirmed that a curious phenomenon of post-transcriptional silencing, initially observed in plants48, was mediated by fragments of dsRNA which were complementary to the messenger RNA of the silenced genes, and they observed that small quantities of dsRNA can have very significant inhibitory effects49. Three years later, in 2001, Elbashir et al showed that the principle component in the interference phenomenon is a small, 21-nucleotide fragment of dsRNA and they presented data to confirm that these small inhibitor fragments of RNA (small interference RNA or siRNA) are active not only in plant and invertebrate cells, but also in superior vertebrates and humans50,51. This discovery opens up a totally new field in our understanding of gene regulation and, at the same time, affords us a glimpse of a technique with huge potential.
Unlike IFN, RNAi is an exquisitely specific process which acts selectively upon RNA for which it is exactly complementary. In addition, the fragments of small dsRNA (< 30 nucleotides) which are interference mediators do not seem to induce the activation machinery of IFN.
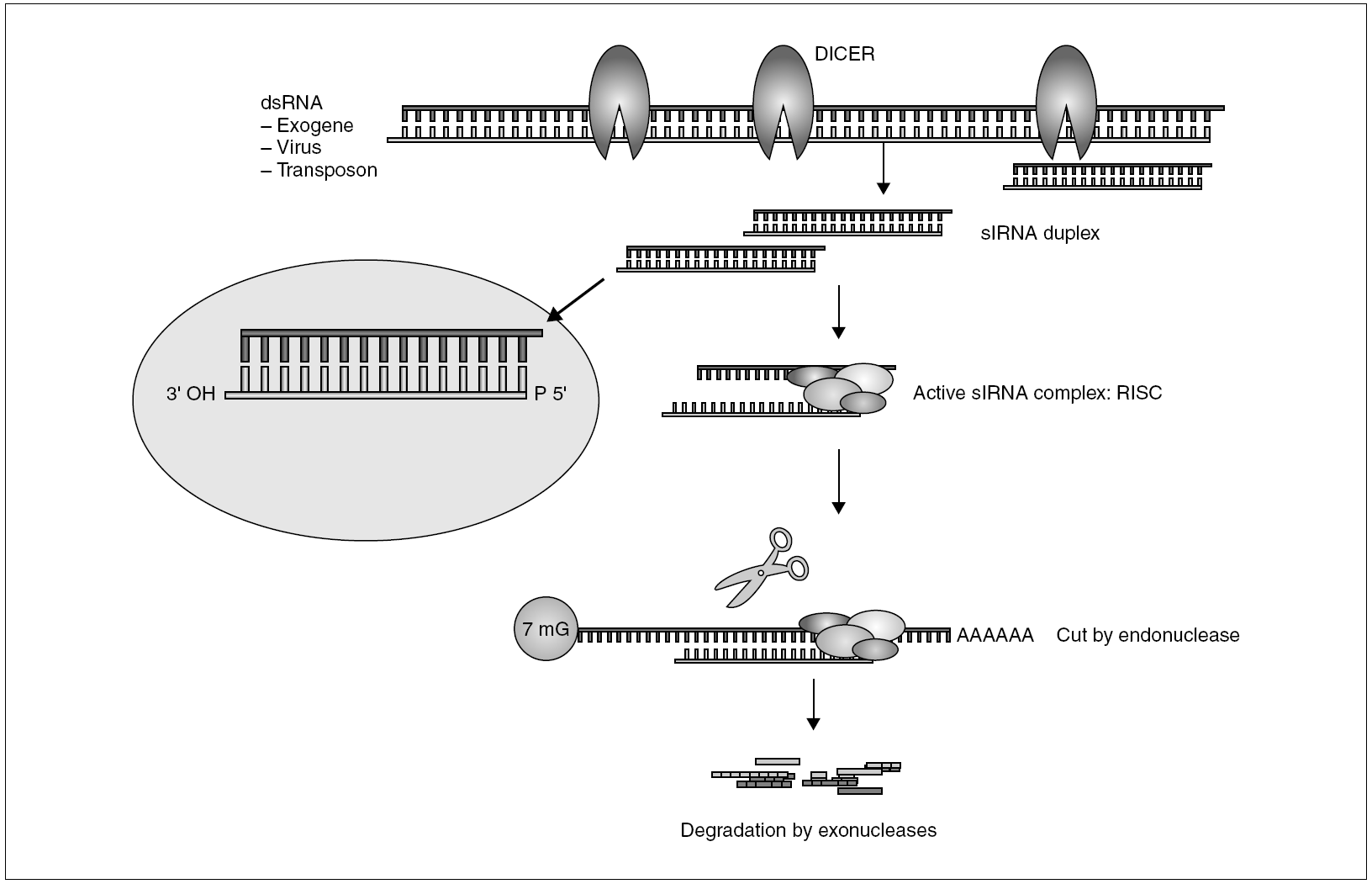
The general mechanism of the production of RNA-mediated interference is shown in figure 2. The presence of intracellular dsRNA activates a specific dsRNA endonuclease known as DICER. This short enzyme cuts dsRNA into 21-23-nucleotide fragments with free 3' ends which constitutes the authentic mediator of the sequence-specific silencing process52. The siRNA fragments then form an active complex with a series of nucleases and helicases known as the RISC (RNA-induced silencing complex)53. During assembly of the RISC, probably only one of the strands is capable of triggering interference54. Using this strand as a guide, the RISC binds to homologous mRNA and catalyzes the cut of the sequence by an RNAse other than DICER, at a short distance from the binding site55. The RNA fragments are then degraded by cellular exonucleases, thus completing the silencing process. In some organisms, C. elegans and plants, the existence has been proposed of a silencing amplification cycle which uses DICER-generated secondary siRNA, on new dsRNA synthesized by an RNA-dependent RNA polymerase (RdRP). This RdRP uses mRNA as a mold and one of the strands of the siRNA fragments as a primer56. This amplification mechanism has been associated with the systemic response exhibited by RNAi in plants and C. elegans, where silencing extends to far zones and can even be transmitted to the progeny49,57. This systemic effect has been confirmed in vertebrates, probably due to the absence of RdRP. The RNA-mediated interference phenomenon is common to most eukariotic cells (it does not exist in bacteria) and seems to be a former defense mechanism which precedes the evolutionary separation between animals and plants. Nevertheless, the basic interference mechanism has gradually developed in different directions. In plants, it fulfills a basically defensive function against virus and viroids; in superior vertebrates and humans it seems to be more a regulator of gene expression. This was revealed with the discovery of genes whose product was not a protein, but microRNA (miRNA)58, short fragments of RNA with interesting regulating properties.
Figure 2. General mechanism of RNAi.
Silencing machinery seems to be conserved in mammals, as indicated in silencing experiments when synthetic siRNA is introduced into the cell, although we do not have proof that the interference phenomenon plays an important antiviral role in natural infection, as is the case with plants. It is possible that adaptive immunity based on the recognition of protein structures has been able to supplant the interference mechanism. It is just as striking that RNAi functions as a defense mechanism in plants and invertebrates, which, in fact, lack adaptive immunity.
Applications of RNAi. How is it used?
The simplest way to introduce RNAi into a cell is by a relatively simple transfection procedure using synthetic RNA. Most of the experiments carried out to date have used this strategy with cells in culture and have obtained consistent and reproducible results. It is also possible to introduce synthetic RNA duplex directly into some living organisms such as C. elegans59,60, in which the interference phenomenon is achieved by "ingestion", simply by adding the siRNA to the culture medium49. In vertebrates, this process seems to be somewhat more complicated: by using large quantities of synthetic RNA, it is possible to introduce siRNA into mice intravenously by a system of hydrodynamic injection61,62. siRNA applied by this mechanism in mice rapidly finds its way to the liver cells and seems to have a surprising stability for at least 7-10 days62. The disadvantages of this strategy of direct administration are the high cost of synthetic RNA (unlike DNA), and particularly, the duration of silencing which, logically, is transitory while the presence of intracellular RNA duplex is maintained. An alternative method of obtaining greater stability is using one of the genetic vectors in the form of plasmids designed to express double-stranded RNA. Lastly, it has been shown that it is possible to generate vectors based on recombinant lentiviruses and retroviruses which can integrate a cassette expressing siRNA into the target cell63-65. This system, by integrating the provirus into the cell genome, allows stable expression of siRNA and would therefore be one of the most attractive for therapy.
Therapeutic applications of RNA interference. The proof of concept
One of the most spectacular applications of RNAi has been that by the Judy Lieberman group in a murine model of Fas-induced fulminant hepatitis62. The administration of an agonist antibody of Fas causes death in mice by massive Fas-dependent hepatic apoptosis in 2-3 days. Using synthetic siRNA and a hydrodynamic intravenous injection system, which leads to a fleeting increase in blood pressure66,67 they managed to protect the mice which had been injected with specific siRNA of the Fas messenger RNA. The rats which had been injected with unspecific siRNA, or modifications of the Fas siRNA, were not protected, thus showing the specificity of the process. The proof of concept that RNAi can cure seems to be well established, although there are still important reservations about the quantities of synthetic RNA that would be necessary, and the possible application of the hydrodynamic system in humans, which would require the intravenous infusion of more than one liter of fluid in a few seconds66,67.
The immune system of the genome. RNAi as an antiviral
With regard to the possible role of RNA interference as an antiviral in humans, there are still some areas which must be better understood in order to evaluate the future of this tool. Is siRNA produced during a natural infection? Despite knowing that, as we have observed, the development of RNA interference in plants is evolutionary, most likely as a defense mechanism against viruses and transposons, we have yet to prove that interference RNA in vertebrates forms part of the natural response to viral infection68. Expression of DICER appears to be very low in differentiated cells and only in some embryonic cells are high levels of DICER detected. Therefore, the natural antiviral function of RNAi would be conserved69. Furthermore, it has not been possible to identify viral genes whose function is to avoid the action of RNAi, in contrast with the numerous examples of viruses which devote many of their genes to hindering a suitable response to interferon, presentation of antigens or adaptive immune response70-73. Nevertheless, silencing machinery is conserved and the introduction of RNAi by different techniques has proven to be very effective in different in vitro models of viral infection, including the hepatitis C virus74-77, hepatitis B virus78, papilloma virus79, polio virus80, rotavirus81 and other RNA viruses82.
Although there is great enthusiasm for the use of RNAi as an antiviral, it is not problem-free. The extraordinary specificity of RNA interference which demands an almost perfect homology between the 21-23 nucleotides which establish the silencing guide, can also be a weak point when it is applied as an antiviral tool. We have only to consider Gitlin et al80, who, while working with a model of RNA interference for poliovirus detected a variant with the ability to escape from the interference mechanism. This variant presented a single substitution in the central target zone of the RNAi.
RNA interference in HIV infection
HIV has been one of the logical first objectives for the application of RNAi technology. Not in vain is it one of the infections in which we have a more precise knowledge of the molecular and cellular processes involved in the process. In the case of HIV, there are data which indicate that genomic RNA can be inactivated by RNAi before integration83 and it has even been possible to achieve significant reductions in HIV RNA before the first hour post-infection in cells transfected with synthetic siRNA against vif and nef regions84. Preventing the HIV genome from entering the nucleus of the cell is extremely important, and this must be fully studied in more stable infection models using primary isolations and cells.
Theoretically, the same active RNAi constructions against genomic RNA would be active against the corresponding mRNAs which encode the structural and accessory proteins of HIV. Different studies confirm the usefulness of RNA interference in blocking the messenger of the accessory proteins Rev85, Tat83, Vif84 and structural proteins, mainly CA86,87. If, as it appears, RNA interference is capable of binding and destroying both the genomic RNA of HIV and the mRNA produced by the integrated provirus, we would be faced with a unique method with activity against the early and late phases of infection84,88.
Lastly, it has been shown that RNA interference can be applied to prevent the expression of virus receptor molecules in the cell membrane. By using synthetic siRNA, a significant effect has been shown on infection by blocking the expression of CD487, CXCR4 and CCR586. The use of interference RNA against cellular products could be advantageous, as suggested by an interesting study which examined the stability of synthetic siRNA in human macrophages. The siRNA of p24 had a half-life of under 7 days in non-infected cells, compared with the more than 15 days of persistence of that which targeted CCR586, whereas in infected cells it was comparable, which suggests that the presence of target RNA may be necessary for the stability of siRNA.
The usefulness of RNA interference seems to be equally confirmed in more physiological models using primary cells86,89 and in stable models for the maintained expression of siRNA86. This last study used a system of transduction with a recombinant lentivirus expressing anti-CCR5 siRNA in primary lymphocytes in which it was possible to reduce the expression of CCR5 by a factor of 10, leading to a reduction in HIV infection by a factor of 3-790. Similarly, encouraging results have been reported concerning the stable inhibition of Tat and expression of CCR5 in primary macrophages using a lentiviral vector91.
We are well aware of the extraordinary capacity of HIV to generate variability and escape from the selective pressure induced by the immune response92,93 and antiretroviral drugs94, including those designed to antagonize the receptor function95,96. It is not surprising that there have been reports of mutants which can escape from the action of siRNA, in this case by a variant of HIV which presented an ad hoc mutation in the 21 nucleotides target zone of the RNAi, corresponding to the tat region. It will therefore be necessary, if we are to guarantee the success of the therapeutic application of RNA interference in HIV infection, to use the combination of different viral and cellular factors.
Our capacity to cure, or to contribute to curing, a disease such as aids using molecular methods will depend on the availability of an efficacious tool, and RNAi seems to fit the bill, but it will also be necessary to update systems to make RNAi reach susceptible cells efficaciously and safely. Today, we are more aware of the possibilities and limitations of a field which can only advance on the basis of scientific knowledge and the subsequent search for solutions to current problems.