The aim of this study was to describe the evaluation of the use of MALDI-TOF MS for the identification of non-tuberculous mycobacteria (NTM) and Mycobacterium tuberculosis directly from liquid MGIT cultures from January 2017 to December 2017.
Material/methodsA total of 155 isolates (mainly respiratory) were analyzed by MALDI-TOF MS (Bruker Daltonics) directly from MGIT liquid medium with a previous extraction procedure.
ResultsMALDI-TOF MS generated acceptable scores for 152 isolates (98.06%). Fifty isolates were identified as M. tuberculosis complex and the remaining 105 as NTM (M. abscessus subsp. abscessus, M. avium, M. celatum, M chelonae, M. chimaera, M. fortuitum, M. gordonae, M. intracellulare, M. kansasii, M. lentiflavum, M. mageritense, M. mucogenicum and M. xenopi).
ConclusionsThese results indicate that MALDI-TOF MS can be useful to identify mycobacteria directly from MGIT cultures and is an accurate, rapid and cost-effective system to be used as a routine method.
Evaluamos la espectrometría de masas (MALDI-TOF MS [Bruker Daltonics]) para la identificación de micobacterias no tuberculosas (MNT) y Mycobacterium tuberculosis a partir de cultivos líquidos (MGIT) desde enero del 2017 a diciembre del 2017.
MétodosSe analizaron mediante MALDI-TOF MS 155 cultivos MGIT positivos, principalmente de origen respiratorio. Previamente a la realización de MALDI-TOF se realizó un procedimiento de extracción directamente del MGIT.
ResultadosMediante MALDI-TOF MS se identificó correctamente a partir del MGIT el 98,06% (n=152) de los aislados. Cincuenta aislados se identificaron como M. tuberculosis complex y los 105 restantes como MNT (M. abscessus subsp. abscessus, M. avium, M. celatum, M chelonae, M. chimaera, M. fortuitum, M. gordonae, M. intracellulare, M. kansasii, M. lentiflavum, M. mageritense, M. mucogenicum y M. xenopi).
ConclusionesEstos resultados indican que MALDI-TOF es una técnica precisa, rápida y coste-efectiva para identificar micobacterias directamente a partir de medios de cultivo líquidos en la rutina diaria.
The genus Mycobacterium includes obligate pathogens, opportunistic pathogens, and saprophytes. Close to 200 mycobacterial species have now been described, and their number is increasing steadily.1 Common methods for the identification of mycobacteria were classically based on biochemical tests. They are slow, laborious and with little capacity to discriminate the multiple mycobacterial species described today. Molecular methods including PCR-based hybridization and sequencing methods are the most used, but have a high cost.2,3 The microscopy study and culture of samples continues crucial for diagnosing mycobacterial infections. Bactec MGIT 960 is a fully automated system, which provides continuous monitoring to recognize positive cultures and thus decrease the response time.4,5
Matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) has been introduced in the laboratories as a powerful tool for the routine identification of bacteria, yeast and moulds. The identification is based on species-specific spectral fingerprints obtained from crude extracts of whole cells.6,7 The speed, robustness and comparatively low costs of simple preparation and measurement make it exceptionally well suited for routine and high through put use. It reduces turnaround time and may potentially impact on benefiting patients.
There are reports for a reliable identification of mycobacteria from solid culture media such the Lowenstein-Jensen or Middlebrook 7H10 or 7H11 medium by the use of MALDI-TOF MS.8,9
The aim of this study is to show a relatively easy method to get identification of mycobacteria from MGIT liquid medium with reliable and reproducible results.
Material and methodsBacteria, media and growth conditions. 224 consecutive positive isolates of clinical samples (mainly respiratory) were analyzed. All samples submitted for mycobacterial cultures were inoculated into the MGIT broth, cultured into Bactec MGIT 960 instrument (Becton Dickinson™) and incubated at 35°C.
Procedure. To work with viable mycobacteria, biosafety level 3 is required. However, the MALDI-TOF used for routine identification is located outside this room. Therefore, we first developed a method to inactivate the mycobacteria before placing them on the target plate. When a MGIT tube was detected as positive, 1mL of the culture from the bottom of the tube was transferred to a 1.5mL eppendorf reaction tube. The eppendorf tube was inactivated for 30min at 95°C (thermoblock) and then centrifuged at 13,000–15,000rpm for 15min, after that, the supernatant was completely removed by a pipette. The bacterial pellet was re-suspended with 300μL of water (HPLC) and vortexed for 2min. Then it was mixed with 900μL of ethanol (100%) and vortexed for 2min. The suspension was centrifuged at 13,000–15,000rpm for 5min and then the supernatant was completely removed. The sediment was allowed to dry with the tube open (∼2–5min), and heated a few minutes if necessary, since it is very important that the sediment is well dry. The remaining liquid was removed with a Pasteur pipette and silica beads (1.5mm zirconite/silica beads), approximately 2 or 4 beads, and 20μL of 100% acetonitrile were added, mixed well with a pipette (undoing the pellet), and then vortexed 2–4min. After waiting for a minute, the pellet was mixed with 20μL 70% formic acid (same volume as acetonitrile), mixed again very well with the pipette until undoing the pellet, and then vortexed another 2–4min. After waiting for a minute, the suspension was centrifuged at 14,000rpm for 2min. Two microliter of the supernatant was spotted onto a MALDI-TOF MS target plate and allowed to air dry. After well dried, 2 additional μL were added and allowed to air dry again. Then 2μL of the matrix solution were added and allowed to air dry. The target plate was run into the MALDI-TOF MS (Bruker Daltonics™) to obtain the identification.
When the results obtained were not satisfactory, the tubes were frozen at −70°C and the next day immediately after taking them out, they were vortexed for 2min, spotted 2μL of the mixture onto a MALDI-TOF MS target plate and air dried. Then, 2μL of the matrix solution were added and allowed to air dry. In case of unsatisfactory results (score<1.6), GenoType Mycobacterium AS/CM/MTBC® was performed. Identification results with score <1.6 were not accepted.
Data analysis. MALDI-TOF MS Analysis. The target plate prepared before (as described before) is inserted into the MALDI-TOF MS (Bruker Daltonics™) for analysis. A composite profile of proteins with a mass-to-charge ratio (m/z) of 2000 to 20,000 is generated based on a minimum of 240 measurements (laser shots) for each analyzed. The composite profile is analyzed using MALDI Biotyper (Mycobacteria Library 4.0). Scores>2.0 are considered suitable, and scores between 1.6 and 2.0 are considered consistent if the same identification is repeated in most of the 10 possibilities provided.
ResultsBefore the implementation of the procedure, a comparative study was performed between MALDI-TOF and GenoType Mycobacterium CM/AS (Hain Lifescience GmbH®). The results are shown in the supplementary material section (Supplementary Table 1).
A total of 5622 clinical specimens were submitted to our laboratory from January 2017 to December 2017 and cultured in liquid MGIT (BACTEC 960). MGIT cultures were positive for mycobacteria in 224 specimens.
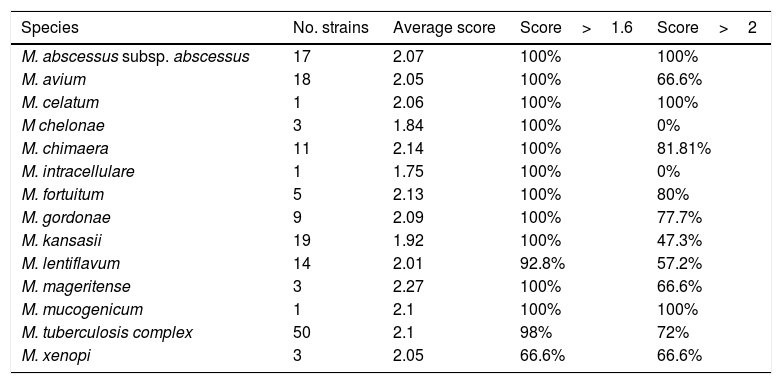
Among them, 69 were identified directly as positive for M. tuberculosis by the chromatographic immunoassay BD MGIT™ TBc Identification Test. In these isolates the identification by MALDI-TOF was not carried out. The remaining 155 MGIT broths were identified by MALDI-TOF MS as described before. The results are shown in Table 1.
Rates of correct identifications of different mycobacterial species by MALDI-TOF MS.
| Species | No. strains | Average score | Score>1.6 | Score>2 |
|---|---|---|---|---|
| M. abscessus subsp. abscessus | 17 | 2.07 | 100% | 100% |
| M. avium | 18 | 2.05 | 100% | 66.6% |
| M. celatum | 1 | 2.06 | 100% | 100% |
| M chelonae | 3 | 1.84 | 100% | 0% |
| M. chimaera | 11 | 2.14 | 100% | 81.81% |
| M. intracellulare | 1 | 1.75 | 100% | 0% |
| M. fortuitum | 5 | 2.13 | 100% | 80% |
| M. gordonae | 9 | 2.09 | 100% | 77.7% |
| M. kansasii | 19 | 1.92 | 100% | 47.3% |
| M. lentiflavum | 14 | 2.01 | 92.8% | 57.2% |
| M. mageritense | 3 | 2.27 | 100% | 66.6% |
| M. mucogenicum | 1 | 2.1 | 100% | 100% |
| M. tuberculosis complex | 50 | 2.1 | 98% | 72% |
| M. xenopi | 3 | 2.05 | 66.6% | 66.6% |
Direct identification detected 50 MGIT broths positive for M. tuberculosis and the remaining 105 were NTM.
MALDI-TOF MS generated acceptable confidence scores (score>2.000 or >1.600) if the same species was repeated in the 10 possibilities provided by the system for 152 (98.06%). In one culture, no identification was obtained due to the poor quality of the spectra. This sample was sent to a reference laboratory and was identified as M. lentiflavum.
Scores obtained for the more frequently isolated species were M.abscessus (100%>2.0, 100%>1.6), M. avium (66.6%>2.0, 100%>1.6), M. kansasii (47.3%>2.0, 100%>1.6) and M. tuberculosis complex (72%>2.0, 98%>1.6).
DiscussionThe usefulness of MALDI-TOF for the identification of mycobacteria has been demonstrated, but there are multiple aspects that complicate and intervene in the results obtained in the identification of mycobacteria by MALDI TOF MS. These factors are the inactivation method of microorganisms, the extraction of the protein material, the type of culture medium and the incubation time.10
Some studies have demonstrated the ability of MALDI-TOF MS for mycobacterial identification from solid media as Saleeb et al.8 and Mediavilla-Gradolph et al.11 Their results were similar to ours but using a more time-consuming method because of need to subculture a positive MGIT tube onto Middlebrook 7H11 agar or Lowestein-Jensen agar. They also obtained poor results with M. abscessus subsp. abscessus and M. abscessus subsp. massiliense which are closely related phylogenetically. Recent reports have demonstrated the ability to identify correctly a large number of mycobacteria from liquid media. The results from the study of Huang et al.12 were similar to ours, but their procedure was different in some aspects. They needed more culture fluid (3ml) and they used ethanol for the inactivation step. In addition, in some cases they needed a washing step with sodium dodecyl sulfate. Miller et al.13 obtained a lower rate of correct identification than in our study since the protein extraction procedure started within 24–72h after the MGIT tube was detected as positive.
In this work, we demonstrate that our procedure for bacterial identification by MALDI-TOF MS is a suitable, reliable, and fast technique for the identification of mycobacteria. Identification can be completed in approximately 90min after the MGIT broths were detected as positive, and only 1ml of culture fluid is used. Our results show that identification of Mycobacterium spp. from a MGIT positive culture is possible using MALDI TOF MS. 152 of the 155 samples that were analyzed by MALDI TOF MS had a successful identification. In general, the identities revealed by the MALDI-TOF MS were consonant with those obtained by other methods.
Therefore, we conclude that it is quite feasible to incorporate MALDI-TOF MS in the clinical microbiology laboratory workflow to identify of mycobacteria from positive MGIT cultures as a routine method, because it is a powerful, rapid and cost-effective method. Identification from liquid cultures can accelerate pathogen identification prior to growth on solid media. Currently in our hospital we have established this procedure as our workflow routine; in addition, this procedure has recently been accredited by ENAC (National Accreditation Entity).
Transparency declarationThis research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.