Sexually transmitted infections (STI) are currently on the increase worldwide. New molecular tools have been developed in the past few years in order to improve their diagnosis. An evaluation was carried out using a new commercially available real-time PCR assay, Anyplex™ II STI-7 (Seegene, Seoul, Korea), which detects seven major pathogens in a single reaction – Chlamydia trachomatis, Neisseria gonorrhoeae, Trichomonas vaginalis, Mycoplasma hominis, Mycoplasma genitalium, Ureaplasma urealyticum, and Ureaplasma parvum – and compared with conventional methods performed in our laboratory.
Materials and methodsTwo different populations were included, and 267 specimens from different sites of infection (urines, endocervical swabs, rectal swabs, vaginal swabs, urethral swabs and one inguinal adenopathy) were processed for both methods.
ResultsThe parameters of clinical performance were calculated for C. trachomatis, N. gonorrhoeae, and T. vaginalis, and the assay achieved sensitivities (SE) from 93.94% to 100%, and specificities (SP) from 96.55% to 100%, with negative predictive values (NPV) from 93.33% to 98.85%, and positive predictive values (PPV) from 96.88% to 100%, with a very good agreement (kappa index from 0.88 to 1).
ConclusionsAnyplex™ II STI-7 is a good tool for the reliable diagnosis of STI. Its ease of use and processing allows it to be incorporated into the day to day laboratory work.
Las infecciones de transmisión sexual (ITS) son actualmente un problema de salud pública en todo el mundo debido al aumento que han experimentado en los últimos años que implica el desarrollo de nuevas herramientas moleculares para mejorar su diagnóstico. Se ha comparado el nuevo ensayo de PCR en tiempo real, Anyplex™ II STI-7 (Seegene, Seúl, Corea) que detecta los siete microorganismos implicados en las ITS – Chlamydia trachomatis, Neisseria gonorrhoeae, Trichomonas vaginalis, Mycoplasma hominis, Mycoplasma genitalium, Ureaplasma urealyticum, and Ureaplasma parvum – en una sola reacción, con los métodos convencionales utilizados en nuestro laboratorio.
MétodosSe incluyeron dos tipos de poblaciones, obteniéndose 267 muestras de diferentes lugares de infección (orines, exudados endocervicales, frotis rectales, frotis vaginales, exudados uretrales y una adenopatía inguinal) que fueron procesadas por ambas metodologías.
ResultadosLas sensibilidades, especificidades y valores predictivos fueron analizados para C. trachomatis, N. gonorrhoeae y T. vaginalis, alcanzando sensibilidades (SE) de 93,94% a 100%, especificidades (SP) de 96,55% a 100%, valor predictivo negativo (NPV) entre 93,33% y el 98,85% y valores predictivos positivos (PPV) de 96.88% a 100% con muy buena correlación (índice kappa de 0.88 a 1).
ConclusionesAnyplexTM II STI-7 es una buena herramienta para el diagnóstico seguro de las ITS. La facilidad de uso y procesamiento permite su incorporación en el trabajo del día a día del laboratorio.
Microbiological diagnosis of syndromes that can be caused by multiple pathogens, such as (STIs) may be challenging. Therefore, methods able to detect multiple microorganisms in a clinical specimen at the same time are essential. The need for the development of reliable, affordable, and effective commercial assays for the management of the syndromic diagnosis of STIs is rising, given their high prevalence and their increasing burden worldwide.1 In 2008, the WHO estimated the total number of incident cases of the four main STIs in adults comprising ages between 15 and 49 years to be 498.9 million. Among them, 105.7 million corresponded to Chlamydia trachomatis, 106.1 million to Neisseria gonorrhoeae, 10.6 million to Treponema pallidum, and 276.4 million to Trichomonas vaginalis.2 More than 30 different bacteria, viruses and parasites can potentially be transmitted through sexual contact, which makes the choice of diagnostic assays a difficult task. A selection of the most relevant pathogens according to local prevalence rates should be targeted.
Conventional diagnostic assays, such as culture and antigen detection assays, lack sensitivity, require viable organisms and thus special shipment conditions and, sometimes, invasive sampling.3 As nucleic acid amplification tests (NAATs) allow us to overcome some of these limitations, several molecular diagnostic assays have recently been commercialized to assist the syndromic diagnosis of STIs. Nowadays, NAATs are flexible and easy to use. In addition, their implicit multiplexing capacity allows for the detection of multiple pathogens in a single sample and, therefore, their implementation in the clinical microbiology laboratory is increasing. The rising prevalence of STIs reported worldwide may be related, at least in part, to the use of improved diagnosis through the use of molecular technologies.2
The detection of multiple pathogens is achieved by NAATs thanks to its ability of multiplexing different assays.4 Several molecular tests are commercially available for the diagnosis of STIs detecting a number of microorganisms ranging from two (i.e. Abbott RealTime™ CT/NG, Abbott Molecular Inc., Des Plaines, IL), to 18 (i.e. CLART® STIs A&B, Genomica S.A.U., Madrid, Spain), thereby simplifying the diagnostic workflow, reducing the hands-on-time, as well as associated costs. Recently, a rapid test that detects Chlamydia trachomatis and Neisseria gonorrhoeae in one step and in 90minutes has been developed for the GeneXpert systems (Cepheid, Sunnyvale, CA). This constitutes the first step toward a highly reliable point-of-care test in the field of STIs.5 Molecular tools have a high sensitivity and specificity for the diagnosis of the most prevalent STIs, and offer the possibility of using non-invasive specimens.6,7
In this study we have evaluated the usefulness of Anyplex™ II STI-7 (Seegene, Seoul, Korea), both for screening and diagnosis of STI. This real-time PCR assay detects seven major pathogens that cause STIs – Chlamydia trachomatis, Neisseria gonorrhoeae, Trichomonas vaginalis, Mycoplasma hominis, Mycoplasma genitalium, Ureaplasma urealyticum, and Ureaplasma parvum – in a single reaction.
Material and methodsStudy designThis was a retrospective cross-sectional observational study designed for the clinical evaluation of the commercial Anyplex™ II STI-7 molecular assay in comparison with conventional diagnostic assays and other molecular assays used for the routine diagnosis of STI at the Microbiology Department of the “Germans Trias i Pujol” University Hospital (HUGTIP, Badalona, Spain). This study was approved by the Ethics Committee at our institution.
Study population and clinical samplesTwo different study populations were included in the present study. Group 1 included 234 individuals attended at the emergency room, the urology and gynecological departments at HUGTIP, primary care centers seeking medical care, as well as young adults (≤25 years old) suspected of having an STI and recruited for C. trachomatis prevalence studies at sexual and reproductive health centers. After routine microbiological diagnosis (culture and Abbott RealTime™ CT/NG), residual sample volume was stored at −20°C until used for the evaluation of the Anyplex™ II STI-7 assay. Group 2 included 33 HIV-negative MSM having high-risk sexual practices that were periodically screened for STIs at the BCN Checkpoint center (a community-based detection center of HIV and other STIs). After being tested by the Abbott RealTime CT/NG PCR Assay, residual sample volume was stored at −20°C until used for this evaluation study.
Clinical specimens included 105 first-void urines, 50 rectal swabs, 18 urethral swabs, and three inguinal lymphadenopathy samples from patients suspected for lymphogranuloma venereum (LGV), 38 endocervical swabs and 53 vaginal swabs. All specimens were anonimized prior to testing.
Conventional microbiological diagnosisIn order to isolate N. gonorrhoeae and T. vaginalis from patients suspected of having an STI, endocervical, urethral and vaginal swabs were cultured onto Chocolate agar PolyViteX (bioMèrieux, Marcy-l’Étoile, France), Chocolate agar PolyViteX VCAT3 (bioMérieux), Columbia agar+5% sheep blood (bioMérieux), Sabouraud dextrose agar (bioMérieux) and Diamond media (Maim SL, Barcelona, Spain). N. gonorrhoeae was identified by biochemical methods (Vitek NH cards, bioMérieux), after 48–72h of incubation in an aerobic atmosphere and T. vaginalis was identified by a wet mount examination after 4–5 days of incubation.
Molecular diagnostic assaysNucleic acid extractionDNA extraction was performed with NucliSENS EasyMAG (bioMérieux), which is an automated system able to extract total nucleic acids from a variety of sample types and volumes. The system automates an enhanced magnetic silica version of BOOM® technology, a very good tool for the universal extraction of RNA and DNA.8
Routine detection of C. trachomatis and N. gonorrhoeaeAll tested specimens were collected with the multi-Collect Specimen Collection Kit (Abbott Molecular Inc.), as recommended by the manufacturer. After nucleic acid extraction, the Abbott RealTime™ CT/NG (Abbott Molecular Inc.) assay was performed according to the manufacturer instructions. This method allows to qualitatively detection of the plasmidic DNA of C. trachomatis and the genomic DNA of N. gonorrhoeae.
Routine C. trachomatis typingAll specimens that were positive for C. trachomatis from patients suspected of having LGV, were subjected to LGV serovar typing by using the Ct-Dt array (Laboratory Biomedical Products BV, Rijswik, The Netherlands).9
Detection of seven STI agents by the novel real-time PCR assayAfter DNA extraction, detection of C. trachomatis, N. gonorrhoeae, T. vaginalis, M. hominis, M. genitalium, U. urealyticum, and U. parvum was performed with the Anyplex™ II STI-7 Detection assay, CE marked since 2012. This multiplex assay uses the newly developed TOCE™ (tagging oligonucleotide cleavage and extension) and DPO™ (dual priming oligonucleotide) technologies10,11. Additionally, semi-quantitative results are obtained based on cyclic-CMTA (catcher melting temperature analysis). The reaction was performed according to the manufacturer in a CFX96 realtime termocycler (bio-Rad, Hercules, CA, USA).
Statistical analysesSensitivity, specificity, positive and negative predictive values (PPV and NPV) of the Anyplex™ II STI-7 Detection assay were calculated were calculated in comparison with the routine diagnostic methods used at our laboratory for the detection of C. trachomatis, N. gonorrhoeae, and T. vaginalis, which were considered as the gold standard with openepi software, www.openepi.com (Emory University, Atlanta, USA). Since Mycoplasma and Ureaplasma infections are not routinely tested, no statistical analyses were performed for these microorganisms.
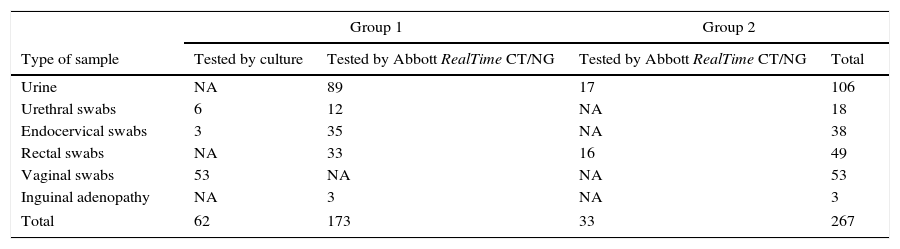
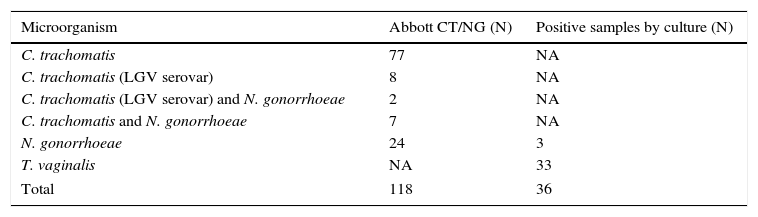
ResultsA total of 267 specimens were tested by the Anyplex™ II STI-7 assay, including positive (n=154, 57.67%) and negative (n=113, 42.32%) specimens according to routine diagnostic assays (culture and/or Abbott RealTime™ CT/NG) and are described in Table 1. Among specimens with a positive result by Abbott RealTime™ CT/NG (n=118), 77 were positive for C. trachomatis and 24 N. gonorrhoeae in single infection, 9 (7.63%) showed a coinfection with C. trachomatis and N. gonorrhoeae. Ten out of 94 (10.63%) C. trachomatis positive specimens were typed as LGV (serovar L2) Table 2.
Specimens tested by the STI-7 assay according to patient group, specimen type and routine diagnosis methods.
| Group 1 | Group 2 | |||
|---|---|---|---|---|
| Type of sample | Tested by culture | Tested by Abbott RealTime CT/NG | Tested by Abbott RealTime CT/NG | Total |
| Urine | NA | 89 | 17 | 106 |
| Urethral swabs | 6 | 12 | NA | 18 |
| Endocervical swabs | 3 | 35 | NA | 38 |
| Rectal swabs | NA | 33 | 16 | 49 |
| Vaginal swabs | 53 | NA | NA | 53 |
| Inguinal adenopathy | NA | 3 | NA | 3 |
| Total | 62 | 173 | 33 | 267 |
NA, not applicable.
Specimens with a positive result by methods used for routine diagnosis.
| Microorganism | Abbott CT/NG (N) | Positive samples by culture (N) |
|---|---|---|
| C. trachomatis | 77 | NA |
| C. trachomatis (LGV serovar) | 8 | NA |
| C. trachomatis (LGV serovar) and N. gonorrhoeae | 2 | NA |
| C. trachomatis and N. gonorrhoeae | 7 | NA |
| N. gonorrhoeae | 24 | 3 |
| T. vaginalis | NA | 33 |
| Total | 118 | 36 |
NA, not applicable.
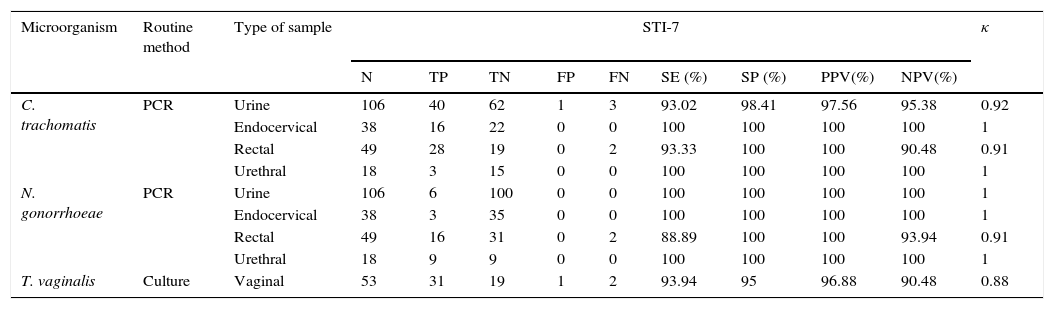
Sensitivity, specificity, PPV, NPV values and kappa coefficient (κ) of the Anyplex™ II STI-7 assay for C. trachomatis, N. gonorrhoeae, and T. vaginalis in comparison with routine diagnostic methods used in our laboratory by type of sample are shown in Table 3. The κ coefficient obtained shows a very high agreement between both methods with κ values from 0.88 to 1 (κ coefficient CI 95%). Out of the 267 specimens, there were 11 discrepancies, 4 urines, 4 rectal swabs and 3 vaginal swabs. All of the vaginal swabs were tested in women who present leucorrhoea and seek medical attention. The rest of the discrepancies corresponded to MSM having high-risk sexual practices, which were asymptomatic or paucisymptomatic.
Performance of STI-7 in comparison with routine methods.
| Microorganism | Routine method | Type of sample | STI-7 | κ | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N | TP | TN | FP | FN | SE (%) | SP (%) | PPV(%) | NPV(%) | ||||
| C. trachomatis | PCR | Urine | 106 | 40 | 62 | 1 | 3 | 93.02 | 98.41 | 97.56 | 95.38 | 0.92 |
| Endocervical | 38 | 16 | 22 | 0 | 0 | 100 | 100 | 100 | 100 | 1 | ||
| Rectal | 49 | 28 | 19 | 0 | 2 | 93.33 | 100 | 100 | 90.48 | 0.91 | ||
| Urethral | 18 | 3 | 15 | 0 | 0 | 100 | 100 | 100 | 100 | 1 | ||
| N. gonorrhoeae | PCR | Urine | 106 | 6 | 100 | 0 | 0 | 100 | 100 | 100 | 100 | 1 |
| Endocervical | 38 | 3 | 35 | 0 | 0 | 100 | 100 | 100 | 100 | 1 | ||
| Rectal | 49 | 16 | 31 | 0 | 2 | 88.89 | 100 | 100 | 93.94 | 0.91 | ||
| Urethral | 18 | 9 | 9 | 0 | 0 | 100 | 100 | 100 | 100 | 1 | ||
| T. vaginalis | Culture | Vaginal | 53 | 31 | 19 | 1 | 2 | 93.94 | 95 | 96.88 | 90.48 | 0.88 |
TP, true positives; TN, true negatives; FP, false positives; FN, false negatives; SE, sensitivity; SP, specificity; PPV, positive predictive value; NPV, negative predictive value, κ, Kappa coefficient.
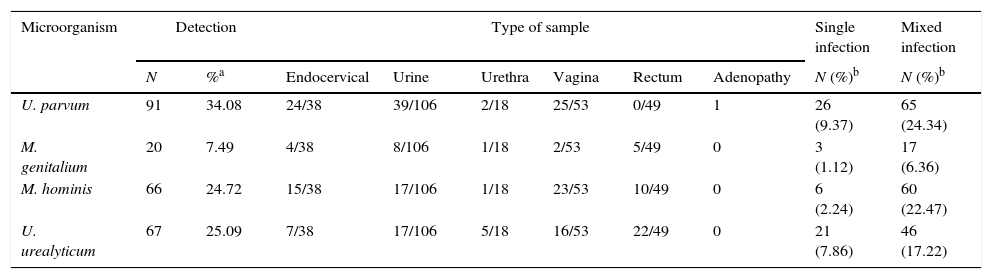
Anyplex™ II STI-7 additionally detected M. hominis, M. genitalium, U. parvum or U. urealyticum in 162 (60.67%) out of 267 specimens either in single or in mixed infections. U. parvum was the most frequent detected microorganism (34.08%). A frequent combination that it was already described was the association between U. parvum and C. trachomatis (Table 4).
Distribution of genital ureaplasma and mycoplasma by specimen.
| Microorganism | Detection | Type of sample | Single infection | Mixed infection | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| N | %a | Endocervical | Urine | Urethra | Vagina | Rectum | Adenopathy | N (%)b | N (%)b | |
| U. parvum | 91 | 34.08 | 24/38 | 39/106 | 2/18 | 25/53 | 0/49 | 1 | 26 (9.37) | 65 (24.34) |
| M. genitalium | 20 | 7.49 | 4/38 | 8/106 | 1/18 | 2/53 | 5/49 | 0 | 3 (1.12) | 17 (6.36) |
| M. hominis | 66 | 24.72 | 15/38 | 17/106 | 1/18 | 23/53 | 10/49 | 0 | 6 (2.24) | 60 (22.47) |
| U. urealyticum | 67 | 25.09 | 7/38 | 17/106 | 5/18 | 16/53 | 22/49 | 0 | 21 (7.86) | 46 (17.22) |
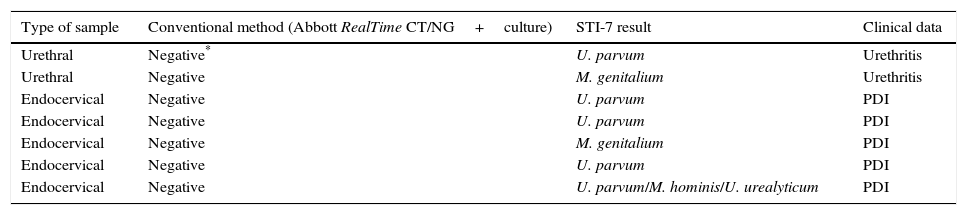
One or more of these four microorganisms were detected in 59 (52.21%) out of 113 specimens that were negative by the routine diagnostic assays used but in only 7 cases had been considered with clinical significance (Table 5).
Characteristics of subjects with Ureaplasma or Mycoplasma in single infection with clinical significance (in the absence of other STI agents).
| Type of sample | Conventional method (Abbott RealTime CT/NG+culture) | STI-7 result | Clinical data |
|---|---|---|---|
| Urethral | Negative* | U. parvum | Urethritis |
| Urethral | Negative | M. genitalium | Urethritis |
| Endocervical | Negative | U. parvum | PDI |
| Endocervical | Negative | U. parvum | PDI |
| Endocervical | Negative | M. genitalium | PDI |
| Endocervical | Negative | U. parvum | PDI |
| Endocervical | Negative | U. parvum/M. hominis/U. urealyticum | PDI |
Overall, the assay was easy to perform and the results generated by the software are easy to interpret, leading to diagnosis in 3hours.
DiscussionThe present study aimed to evaluate in the clinical setting a molecular real-time PCR-based assay (Anyplex™ II STI-7) for the diagnosis of STIs in comparison with the reference methods used at our institution. The detection of seven different pathogens from a variety of clinical specimens in a single reaction is an important advantage offered by this molecular method for the differential diagnosis of many common syndromes, such as urethritis and cervicitis. In addition, microorganisms usually not detected by conventional diagnostic methods can be identified by this assay, thus reducing the cases in which a microbiological diagnosis cannot be achieved for a given STI.
C. trachomatis and N. gonorrhoeae are major causative agent of urethritis and cervicitis.12 Given the difficulty in growing this microorganism in culture, over the last years classical microbiological methods have been replaced by NAATs in many laboratories.13 In our study, the Anyplex™ II STI-7 assay detected C. trachomatis with 94.6% sensitivity and 99.2% specificity, and N. gonorrhoeae with 93.8% sensitivity and 100% specificity in comparison with the Abbott RealTime™ CT/NG. Thus, a low proportion of discrepant results were obtained for C. trachomatis in urine specimens (one false-positive and two false-negatives) and in rectal swabs (two false negatives), and for N. gonorrhoeae (two false-negatives), while endocervical and urethral specimens lead to consistent results. Rectal swabs constitute a challenging clinical specimen and, in fact, this sample type is not included in the Abbott RealTime™ CT/NG CE mark, although several studies have demonstrated its usefulness.14,15 Urine is commonly used as an alternative, easier to collect specimen, but may have a lower diagnostic yield in comparison with endocervical or urethral specimens, which are the preferred sample type for the diagnosis of C. trachomatis and N. gonorrhoeae. Additionally, the obtained false-negative results by the STI-7 assay could be explained in part by a potentially lower DNA integrity due to an additional freeze–thaw cycle (specimens were first tested by the Abbott assay and then frozen until tested by the Anyplex™ II STI-7 assay). On the other hand, we cannot completely rule out the possibility of a false-positive result by the Abbott assay in these seven cases. However, this possibility is very unlikely in real-time PCR assays where no post-PCR processing is necessary, and the negative control included in each run was always negative. Regarding the false-positive result (a C. trachomatis detection in a urine sample), the Anyplex™ II STI-7 semi-quantitative result of ++ (amplification detected at a PCR cycle number between 30 and 40) argues against a false-positive result. Thus, specificity could rise to a 100% if this result was considered a true positive, in agreement with a previous study with the prior version of the assay.16 The STI-7 assay was also compared with culture for N. gonorrhoeae, and the overall agreement was good among the 62 specimens tested by both methods, although the number of positive specimens by culture (n=3) was too low to draw definitive conclusions, which is a limitation of our study. Previous studies using molecular methods for the detection of C. trachomatis and N. gonorrhoeae have showed a good agreement with other molecular methods or classic culture for N. gonorrhoeae.4,17,18 In addition, these studies have demonstrated that molecular tools can increase the detection rate of these pathogens in samples from different origins (rectal, vaginal, pharyngeal, urine), and even reduce the number of false positive results due to the presence of flora belonging to different species of Neisseria.16,19 Additionally, a previous study showed a good correlation between this assay and an in-house molecular assay for the detection of N. gonorrhoeae in extragenital sites.14 Therefore, although the studied population was heterogeneous, there was a good agreement between the two molecular methods used both in the MSM group and in group 1 (including hospital departments, primary care, and sexual and reproductive health centers). Consequently, this multiplex PCR could be useful for diagnosing STIs in symptomatic patients as well as for the screening of STIs in selected populations at risk. The microscopic observation of T. vaginalis after culture is subjective, leading to a variable sensitivity in comparison to other diagnostic methods such as molecular tools. Thus, the inclusion of this protozoan parasite in the Anyplex™ II STI-7 assay can provide useful information to improve the syndromic diagnosis of STIs. In our study, sensitivity (93.94%) and specificity (96.55%) of the Anyplex™ II STI-7 test in comparison with culture was good. Importantly, T. vaginalis was detected by this assay and considered clinically relevant (one urine specimen, one endocervical swab and one inguinal adenopathy) that had not been previously tested by culture, since there was no clinical suspicion of an infection by this pathogen. Regarding the limphadenopathy, T. vaginalis was also detected by culture in a vaginal swab from the same patient.
The results obtained for Mycoplasma and Ureaplasma are difficult to interpret, as we did not search for them actively with conventional diagnostic methods, which is a limitation of our study. It should be taken into consideration that these microorganisms were mostly detected in mixed infections rather than in single infection, as reported in a previous study using the Anyplex™ II STI-7 assay.20 Besides, they were more prevalent in urine and vaginal swabs than in the other sample types. A positive PCR test for at least one of these microorganisms was obtained in 59 out of 113 patients with a negative result for the most relevant pathogens (C. trachomatis, N. gonorrhoeae and T. vaginalis). Among them, only in seven cases (12.06%) the detected microorganism was considered clinically relevant (Table 5). Given that Ureaplasma spp. and M. hominis are part of the flora from the genitourinary tract of healthy and sexually active people,21 a follow-up specimen might be useful to confirm their pathogenic role. The identification of these potentially pathogenic microorganisms is highly relevant given that the first-line treatment may not be appropriate for Ureaplasma and Mycoplasma infections. Nowadays it is believed that Mycoplasma plays a significant role in genitourinary tract pathologies and constitutes a risk factor for the development of nongonococcal urethritis and acquisition of HIV infection.22,23 In the study population, U. parvum showed the highest detection rates (34.08%), followed by U. urealyticum with (25.09%). Despite the controversial role played by Ureaplasma and Mycoplasma, Horner et al.23 conclude that U. urealyticum and M. genitalium were associated with 15–25% of both chronic and acute cases of non-gonococcal urethritis. Additionally, M. genitalium has been studied as a cause of urethritis and cervicitis after a first-line treatment relapse24 due to their prevalence and resistance to macrolide treatments. NAAT is the recommended method to detect M. genitalium,1 and the Anyplex™ II STI-7 assay could be a good choice to detect it.
Overall, the present study demonstrates that the Anyplex™ II STI-7 Detection assay is a simple and reliable method for the diagnosis of STIs able to improve the diagnostic methods used in our center. In addition, its multi-sample option and multiplex detection ability is strongly recommended for the detection of multi-etiological syndromes such as STIs, even in asymptomatic carriers and low prevalence populations. Although the detection of U. urealyticum, U. parvum and M. hominis must be carefully interpreted in the clinical setting, especially in non-genital samples such as urine or rectal swabs, this assay can be a useful tool for diagnostic screening programs and also for epidemiological studies in Public Health.
Conflict of interest statementThe authors declare no conflict of interest.
EM holds a Miguel Servet contract (MS09/00044) funded by ISCIII (Spanish Government). This study was partially funded by Werfen, S.A., distributor of Seegene in Spain. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. The authors declare that they have no conflict of interests