The effectiveness of PCR methods to amplify rickettsiae from clinical samples has still not been evaluated. Our aim was to determine the sensitivity and usefulness for Rickettsia species identification by PCR methods, targeting 16S rDNA, htrA, gltA, ompA, and ompB genes for molecular diagnosis of rickettsioses.
MethodsA total of 72 clinical samples (EDTA-blood, skin biopsies and ticks) taken from 52 patients in the early phase of the illness with PCR-confirmed rickettsioses were included. Single [16S rDNA, gltA (5′ end), and htrA genes] and sequential (nested or semi-nested) PCR assays [ompB, gltA (central region) and ompA genes] were performed.
ResultsFor single-stage PCR assays, the greatest sensitivity (33.3%) was obtained using the gltA (5′ end), while for sequential assays, the most sensitive results were obtained using the ompB assay (83.3%). The highest sensitivity (100%) was achieved using the three sequential PCRs. The ompA PCR method was the most reliable for identifying Rickettsia species, according to clinical features.
ConclusionsPCR-based amplification methods are useful rickettsial diagnostic tools in the early phase of the illness. The three sequential PCR assays here investigated (ompB, gltA and ompA) appear to be useful tools for molecular diagnosis of rickettsioses. ompB PCR assay is effective for primary screening, since it detects a high percentage of positive samples. ompA assay is the most useful method to identify a Rickettsia species in human pathology. Nevertheless, epidemiology, clinical symptoms and the vector involved in the infection have to be taken into account for the diagnosis of rickettsioses.
Hasta la fecha no se ha evaluado la eficiencia de ensayos de PCR que amplifican ADN de Rickettsia spp. en muestras clínicas. Nuestro objetivo fue determinar la sensibilidad y la utilidad para la identificación de especies de Rickettsia de métodos de PCR que amplifican los genes ADNr 16S, htrA, gltA, ompA y ompB para el diagnóstico molecular de rickettsiosis.
MétodosEstudiamos 72 muestras (sangre con EDTA, biopsias de piel y garrapatas) de 52 pacientes con rickettsiosis en fase aguda y confirmada por PCR. Se realizaron PCR sencillas [ADNr 16S, gltA (extremo 5’), y htrA] y secuenciales (anidadas o semianidadas) [ompB, gltA (región central) y ompA].
ResultadosLa mayor sensibilidad con PCR sencillas se obtuvo para gltA (extremo5’). En PCR secuenciales, los resultados más sensibles se lograron utilizando ompB (83,3%). La mayor sensibilidad (100%) se obtuvo tras realizar 3 PCR secuenciales. Según las características clínicas, la PCR de ompA fue la más útil para identificación de especies de Rickettsia.
ConclusionesLos métodos de PCR son válidos para el diagnóstico de rickettsiosis en fase aguda. Los 3 ensayos de PCR secuenciales aquí estudiados (ompB, gltA y ompA) son herramientas útiles para el diagnóstico molecular de rickettsiosis. La PCR de ompB es efectiva para un primer cribado y permite detectar un alto porcentaje de muestras positivas. La PCR del gen ompA es la más fiable para implicar una especie de Rickettsia en patología humana. No obstante, la epidemiología, los síntomas clínicos y el vector deben ser valorados para el diagnóstico de rickettsiosis.
Rickettsioses are caused by obligate intracellular bacteria within the genus Rickettsia mainly transmitted by arthropods. Until recently, Mediterranean spotted fever (MSF) caused by R. conorii was considered to be the only tick-borne rickettsiosis (TBR) prevalent in Europe. However, new TBR has been described in Europe during the last years. These new rickettsioses include infections caused by R. sibirica mongolitimonae,1–3R. helvetica,4R. slovaca,5,6R. raoultii,7,8Candidatus R. rioja,7,9R. massiliae,10R. monacensis11 and a MSF-like TBR caused by different subspecies of R. conorii (R. conorii subsp. caspia and R. conorii subsp. israelensis).12,13 In addition, flea-borne rickettsiosis caused by R. felis has been described in Europe.14,15 Furthermore, African tick-bite fever (ATBF), caused by R. africae, is frequently diagnosed in Europe in patients returning from endemic areas.16,17
Clinical symptoms of TBR include fever, headache, muscle pain, rash, local lymphadenopathy and a characteristic inoculation eschar (tache noir) at the site of the bite. However, these clinical signs are not present in all cases and may vary depending on the species implicated. Currently, different laboratory assays which provide a confirmatory diagnosis of these infections are available. Immunofluorescence assay (IFA) is accepted as the reference method but its sensitivity is low in the early stages and cross-reactivity occurs among spotted fever group rickettsiae. Culture must be performed only in Biosafety Level 3 facilities, which are limited to Reference Centers or few research laboratories. Molecular methods based on polymerase chain reaction (PCR) have enabled the development of sensitive, specific and rapid tools to detect rickettsiae in clinical samples, including arthropods. As it has been stated in the European Guidelines for the diagnosis of tick-borne bacterial diseases, ticks themselves can be used as tools for the diagnosis of these diseases in patients.18 Up to date, real-time PCR approaches (more sensitive and ultra-rapid) are not routinely used for molecular diagnosis of rickettsioses since they require expensive equipments and reagents.
Today PCR primer pairs targeting rickettsial genes are widely used. However, the efficiency of previously described PCR assays for rickettsioses has not been compared to one another. The aim of this study was to determine the assay efficiency (sensitivity and validity for the identification of Rickettsia species) of the PCR methods targeting 16S rDNA, gltA, htrA, ompB and ompA that are used in our laboratory routine for the molecular diagnosis of rickettsioses in clinical samples (including ticks removed from patients who had developed illness) chosen according to criteria detailed in ‘Materials and methods’ section.
Materials and methodsFrom a total of 1276 samples received in the Special Pathogens laboratory (a reference lab receiving specimens from patients residing in Spain) at the Hospital San Pedro-Center of Biomedical Research of La Rioja to determine Rickettsia infection from January 2003 to December 2008, we selected 72 clinical samples (body fluids, biopsies and ticks attached to patients) corresponding to 52 patients with PCR-confirmed rickettsioses. Following the European Guidelines for the diagnosis of tick-borne bacterial diseases,18 patients were categorized into groups: DEBONEL/TIBOLA (Dermacentor-borne-necrosis-erythema-lymphadenopathy/Tick-borne-lymphadenopathy), spotted fever patients and patients suffering from fever after tick-bite. It is worth noting that DEBONEL/TIBOLA patients did not suffer from high temperature nor presented a disseminated maculopapular rash.
Samples should follow these criteria: (1) they were collected from patients in the early phase of the illness before antibiotic treatment, (2) they had arrived to our laboratory in optimal conditions, according to the European Guidelines,18 (3) they gave positive PCR results for Rickettsia species with at least one of the PCR methods included in this work (see below). In our Center, according to our experience and the literature, we are constantly updating different PCR methods for improving the molecular diagnosis of patients with rickettsiosis. Thus, in the laboratory we have different PCR tests. For this reason, since the samples that have been processed to carry out this work have come to our laboratory for a period of 5years, these were processed first with the PCR method that was used in the lab at that time, and subsequently, when it was designed this work, each sample that fulfilled the requirements set out above was processed with all PCR methods evaluated in this report.
We made a final diagnosis that considered: (1) epidemiological data; (2) clinical picture, which included clinical syndrome, clinical evolution and response to the therapy; (3) known distribution of Rickettsia spp.; (4) similarity of the nucleotide sequences with a validated Rickettsia species; (5) score of the European Guidelines for the diagnoses of tick-borne bacterial diseases. The establishment of the final diagnosis was always compatible with clinical and epidemiological data. To evaluate the usefulness of the PCR assays for species identification, we analyzed the correspondence among the highest identities of nucleotide sequences with validated rickettsia species for all PCR-positive clinical samples available from the same patient, always taking also into account the clinical syndrome.
The studied samples included EDTA-treated blood (n=16), plasma (n=2), buffy coat (n=4), erythrocyte portions of the specimens (n=12), sera (n=8), skin biopsies (n=3) and ticks (n=27).
Ticks were classified through taxonomic keys19,20 and molecular methods.21
DNA was extracted using commercial kits, according to the manufacturer's instructions: QIAmp DNA Blood minikit (Qiagen, Hilden, Germany) for blood and sera samples and QIAmp DNA Tissue minikit (Qiagen, Hilden, Germany) for skin biopsies and ticks. DNA extracts were used as templates for six PCR assays: three single PCRs [16S rDNA, citrate synthase (gltA; 5′ end), and 17kDa protein (htrA)] and three sequential (nested or semi-nested) PCR assays targeting 120-kDa genus common antigen (ompB), gltA (central region) and 190kDa protein antigen (ompA) gene. Each reaction (50μL) was performed in an automatic thermocycler (Biometra, Bio-Rad) by adding 5μL of 10× PCR buffer (NH4)2SO4, 1.5mM of MgCl2, 0.2mM of dNTPs mix, 1.5U of Taq DNA polymerase (Bioline), 150ng of template DNA, 1μM of each primer and milli-Q water. Details about PCR primer pairs, size of the amplicons (bp), annealing temperatures and type of the assays are shown in Table 1. Two replicates on each specimen were performed for each PCR method. Negative controls (one of them with template DNA but without primers and the other with primers and containing water instead of template DNA) as well as a positive control (R. conorii Malish #7 grown in Vero cells) were included in all assays. Amplicons were directly sequenced in an automatic sequencer (model ABI-PRISM 3130XL, Applied Biosystems, Foster City, CA). Nucleotide sequences were compared with those available at GenBank using Basic Local Alignment Sequence Tool (BLAST) (http://blast.ncbi.nlm.nih.gov/Blast.cgi), and analyzed according to multiple locus sequence typing criteria.29 Sequences were aligned using ClustalW2 program (http://www.ebi.ac.uk/Tools/msa/clustalw2/). Sequences were submitted to National Center for Biotechnology Information (NCBI) GenBank when considered appropriate.
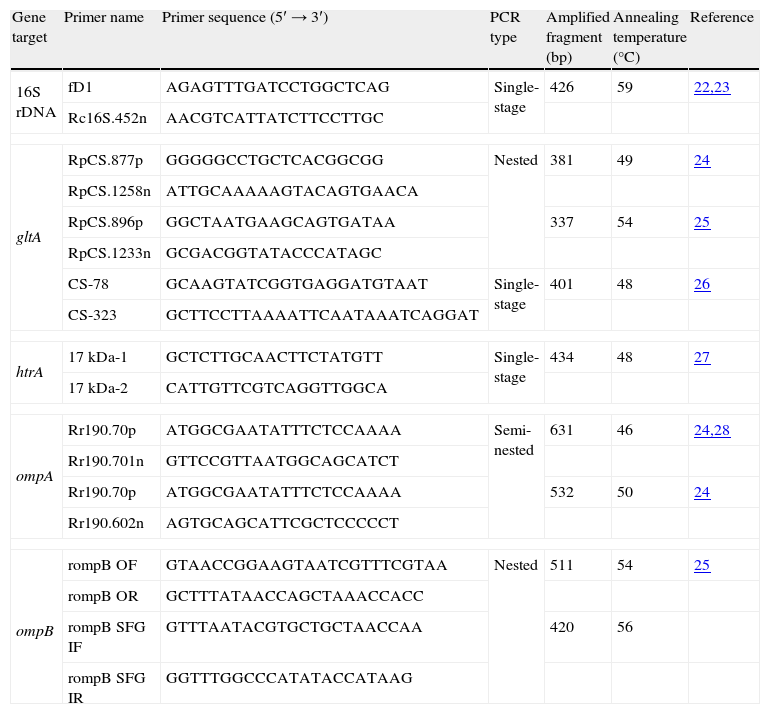
PCR primer pairs used in this study.
| Gene target | Primer name | Primer sequence (5′→3′) | PCR type | Amplified fragment (bp) | Annealing temperature (°C) | Reference |
| 16S rDNA | fD1 | AGAGTTTGATCCTGGCTCAG | Single-stage | 426 | 59 | 22,23 |
| Rc16S.452n | AACGTCATTATCTTCCTTGC | |||||
| gltA | RpCS.877p | GGGGGCCTGCTCACGGCGG | Nested | 381 | 49 | 24 |
| RpCS.1258n | ATTGCAAAAAGTACAGTGAACA | |||||
| RpCS.896p | GGCTAATGAAGCAGTGATAA | 337 | 54 | 25 | ||
| RpCS.1233n | GCGACGGTATACCCATAGC | |||||
| CS-78 | GCAAGTATCGGTGAGGATGTAAT | Single-stage | 401 | 48 | 26 | |
| CS-323 | GCTTCCTTAAAATTCAATAAATCAGGAT | |||||
| htrA | 17kDa-1 | GCTCTTGCAACTTCTATGTT | Single-stage | 434 | 48 | 27 |
| 17kDa-2 | CATTGTTCGTCAGGTTGGCA | |||||
| ompA | Rr190.70p | ATGGCGAATATTTCTCCAAAA | Semi-nested | 631 | 46 | 24,28 |
| Rr190.701n | GTTCCGTTAATGGCAGCATCT | |||||
| Rr190.70p | ATGGCGAATATTTCTCCAAAA | 532 | 50 | 24 | ||
| Rr190.602n | AGTGCAGCATTCGCTCCCCCT | |||||
| ompB | rompB OF | GTAACCGGAAGTAATCGTTTCGTAA | Nested | 511 | 54 | 25 |
| rompB OR | GCTTTATAACCAGCTAAACCACC | |||||
| rompB SFG IF | GTTTAATACGTGCTGCTAACCAA | 420 | 56 | |||
| rompB SFG IR | GGTTTGGCCCATATACCATAAG | |||||
Positive and negative controls gave the expected results in all PCR assays.
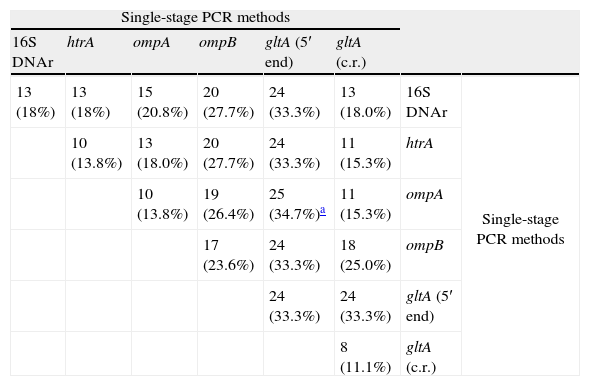
SensitivitySingle-stage PCR assaysSingle-stage PCR assays for gltA (5′ end) allowed the detection of rickettsial DNA in more samples (n=24) than the remaining single-stage PCR tests analyzed. Positive results obtained with each PCR assay are detailed in Table 2. When two single-stage PCR assays were carried out, the greatest sensitivity (25 out of 72; 34.7%) was obtained using gltA (5′ end) and ompA genes. The addition of more single-stage PCRs (3, 4, 5 or 6) targeting analyzed genes did not modify the percentage of positive samples (Table 2).
Number of samples (percentage, %) in which rickettsial DNA was detected using single-stage PCR methods targeting rickettsial genes described.
| Single-stage PCR methods | |||||||
| 16S DNAr | htrA | ompA | ompB | gltA (5′ end) | gltA (c.r.) | ||
| 13 (18%) | 13 (18%) | 15 (20.8%) | 20 (27.7%) | 24 (33.3%) | 13 (18.0%) | 16S DNAr | Single-stage PCR methods |
| 10 (13.8%) | 13 (18.0%) | 20 (27.7%) | 24 (33.3%) | 11 (15.3%) | htrA | ||
| 10 (13.8%) | 19 (26.4%) | 25 (34.7%)a | 11 (15.3%) | ompA | |||
| 17 (23.6%) | 24 (33.3%) | 18 (25.0%) | ompB | ||||
| 24 (33.3%) | 24 (33.3%) | gltA (5′ end) | |||||
| 8 (11.1%) | gltA (c.r.) | ||||||
c.r., central region.
A total of 35 (48.6%) positive samples were obtained using sequential ompA PCR assays. Sequential ompB and gltA (central region) PCR assays, gave positive results for 60 (83.3%) and 42 samples (58.3%) (Table 3).
Number of samples (percentage, %) in which rickettsial DNA was detected using sequential PCR methods targeting rickettsial genes described.
When a sequential PCR assay was combined with each of the single-stage PCRs, the best results (61 out of 72; 84.7%) were achieved with sequential ompB and single-stage ompA PCRs. When adding a third PCR assay (other single-stage one), similar results were obtained for the remaining target genes. On the contrary, the percentages of detection of rickettsial DNA considerably increased when two sequential PCR assays were used: 75% targeting ompA and gltA; 91.6% for ompB and ompA and 94.4% in the case of combining ompB and gltA (Table 3). Rickettsiae were detected in one more clinical sample (95.8%) when single-stage ompA PCR results were added to those obtained from sequential ompB and gltA PCR assays. The three sequential PCR assays targeting ompA, ompB and gltA genes yielded positive results for all studied clinical samples (100%) (Table 3).
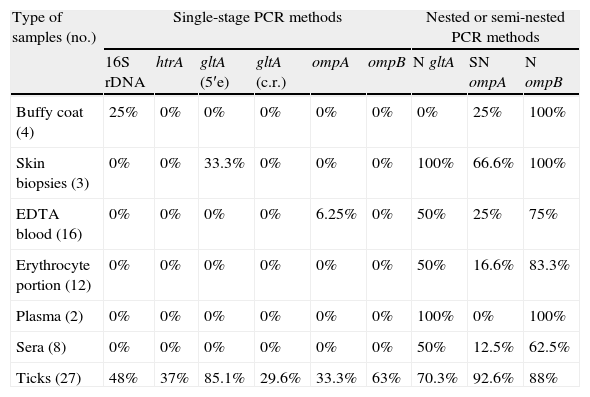
According to the type of sampleSkin biopsies and body fluidsSingle-stage PCR tests targeting gltA (central region) and htrA genes were not useful to detect rickettsial DNA in any of the biopsies and body fluids analyzed. In contrast, rickettsial DNA was found in 100% of skin biopsies and plasma samples using sequential PCRs for gltA and/or ompB. All buffy coat samples gave positive results using sequential ompB PCRs. For EDTA-treated blood samples (n=16), sequential PCR assays for ompA, gltA and ompB allowed the detection of rickettsial DNA from 4, 8 and 12 specimens, respectively. Rickettsiae were detected in 16.6% and 50% erythrocyte portions of specimens using sequential ompA and gltA PCR assays, respectively. For these samples, the best results (83.3%) were obtained using sequential ompB PCR assays. When sequential ompA, gltA and ompB PCR assays were tested with sera specimens, rickettsiae were found for 12.5, 50 and 62.5% specimens (Table 4).
Rate of positive results for 16S rDNA, htrA, gltA, ompA and ompB PCR assays according to the type of sample.
| Type of samples (no.) | Single-stage PCR methods | Nested or semi-nested PCR methods | |||||||
| 16S rDNA | htrA | gltA (5′e) | gltA (c.r.) | ompA | ompB | N gltA | SN ompA | N ompB | |
| Buffy coat (4) | 25% | 0% | 0% | 0% | 0% | 0% | 0% | 25% | 100% |
| Skin biopsies (3) | 0% | 0% | 33.3% | 0% | 0% | 0% | 100% | 66.6% | 100% |
| EDTA blood (16) | 0% | 0% | 0% | 0% | 6.25% | 0% | 50% | 25% | 75% |
| Erythrocyte portion (12) | 0% | 0% | 0% | 0% | 0% | 0% | 50% | 16.6% | 83.3% |
| Plasma (2) | 0% | 0% | 0% | 0% | 0% | 0% | 100% | 0% | 100% |
| Sera (8) | 0% | 0% | 0% | 0% | 0% | 0% | 50% | 12.5% | 62.5% |
| Ticks (27) | 48% | 37% | 85.1% | 29.6% | 33.3% | 63% | 70.3% | 92.6% | 88% |
N, nested PCR; SN, semi-nested PCR; c.r., central region; 5′e, 5′ end.
Single-stage PCR assays for gltA (5′ end), ompB, 16S rDNA, htrA, ompA and gltA (central region) genes showed the presence of rickettsial DNA for 85.1; 63.0; 48.0; 37.0; 33.3 and 29.6% of studied ticks, respectively. Sequential PCR assays for ompA, ompB and gltA genes allowed the detection of rickettsial DNA in 92.6, 88.0 and 70.3% of the ticks, respectively (Table 4).
Detailed results about sensitivity are showed in Tables 2–4.
Usefulness for the identification of Rickettsia speciesBased on clinical features, 26 out of the 52 patients were diagnosed with DEBONEL/TIBOLA. According to 24 ompA nucleotide sequences obtained from EDTA treated blood (n=2) or ticks removed from patients (n=22), there were 9 cases caused by R. slovaca, 14 cases by Candidatus R. rioja and one case by R. raoultii. According to our experience, species identification obtained after sequence analysis of ompA amplicons is accurate since these nucleotide sequences showed 100% correlation with the clinical pictures of the patients. These data were not obtained with the sequencing of other fragment genes analyzed herein. Sequence analysis using other PCR methods did not allow discriminating the rickettsial species implicated or showed highest identity with species that are not present in our area. On the one hand, sequencing of gltA (5′ end) and/or ompB amplicons yielded similar percentages of identity (≥97.7%) with R. raoultii, R. slovaca or Candidatus R. rioja. On the other hand, nucleotide sequences generated for 16S rRNA and htrA genes showed the highest identity (≥99.5%) with those of R. peacockii and R. rickettsii. Partial sequences for 16S rRNA and htrA genes from R. slovaca generated in this study were deposited in GenBank under accession Nos. JQ740394 and JQ740395, respectively.
In the absence of ompA sequences (n=2), data for species identification were not conclusive, and unfortunately, the rickettsial causative agent remained unknown. Nevertheless, DEBONEL/TIBOLA is a well-defined syndrome that can be caused by, at least, three Rickettsia species: R. slovaca, R. raoultii and Candidatus R. rioja. For these two cases, the final diagnosis of DEBONEL/TIBOLA was established according to the PCR positive assays (only considering presence of rickettsiae), joint with the clinical manifestations and the epidemiological features of the patients.
Based on clinical features, 24 patients were diagnosed with spotted fever. In these cases, specimens corresponding to 22 patients gave positive results for sequential ompB PCR assays. When sequenced, the highest identity (≥99.2%) of ompB amplicons corresponded to R. conorii for 10 patients (in 4 of them positive ompA results confirmed this infection) and R. africae for 2 patients. In both cases, detection of R. africae was confirmed by ompA and gltA sequencing (≥99.4% identity). In addition, according to ompB data, R. massiliae was the etiological agent in one spotted fever patient (also confirmed with gltA nucleotide results) and R. felis in 2 patients (confirmed with gltA and ompA sequence analyses). All these patients had clinical and epidemiological data compatible with the molecular diagnosis. Regarding the ticks attached to spotted fever patients (n=3), nucleotide sequences corresponded to R. conorii with all PCR assays studied herein. Similarity percentages were as follows: 100% for ompB and 16S rDNA, ≥99.8% for ompA, 99.7% for gltA (central region) and 99.5% for htrA. Using PCR assays targeting 5′ end gltA, R. conorii and R. raoultii were indistinguishable. In seven patients diagnosed with spotted fever, the rickettsial species remained unknown (Rickettsia sp.) after analysis of ompB and/or gltA nucleotide sequences. Again, the ompA fragment (when available) was the PCR target gene that best correlated with the clinical diagnosis.
Lastly, in two patients with fever after tick-bite, ompB nucleotide sequences corresponded to R. conorii and R. sibirica mongolitimonae, respectively.
DiscussionDetection of rickettsial DNA in clinical samples and arthropods is mainly based, among others, on the amplification of 16S rDNA, htrA, gltA, ompA and ompB genes using conventional PCR assays. The efficiency assay of these PCR targets has not been previously compared to one another, and the quality of the samples has not been taken into account in previous studies. Short term of rickettsiemia in humans, low titers of rickettsiae in human blood and quick elimination following the treatment with doxycycline are intrinsic difficulties for applying PCR assays for the diagnosis of rickettsial diseases. In our work, the sensitivity and usefulness for Rickettsia species identification of these PCR methods in good quality samples that had confirmed-presence of rickettsial DNA was analyzed. It is worth noting that real-time PCR assays are not being routinely used for diagnosis of rickettsioses, thus supporting the importance of regular PCR as recommended in the European Guidelines.18
For single PCR assays, the greatest sensitivity to detect rickettsial DNA in clinical samples (33.3%) was obtained using gltA (5′ end). PCR assays for 16S rDNA and htrA genes were included in our study in order to investigate their sensitivity when testing clinical samples, although it is well-known that these methods have been recommended for the genus level identification of rickettsiae. In addition, PCR primer pairs targeting the same ompA region have been repeatedly used to amplify rickettsiae.30,31
As expected, sequential PCR assays increased the sensitivity. Considerable sensitivity (83.3%) was achieved using sequential ompB PCRs. However, it is well-known that the use of sequential PCR assays is a source of carryover contamination. In our study, we aimed to minimize the risk of contamination. Thus, the work was carried out by experienced people and we used negative, positive and even internal controls, and different rooms for the first and second rounds. The highest sensitivity (100%) was obtained combining three sequential PCR assays (ompA, ompB and gltA). These assays have been used for the molecular diagnosis of rickettsioses,32,33 as well as to identify Rickettsia species in new clinical pictures,1,11,14 and have allowed the genetic characterization of a new Rickettsia (Candidatus R. rioja) implicated in human pathology.9
Our results indicate the usefulness of buffy coat and plasma specimens for molecular detection of rickettsial infection when skin biopsies are not available. Single-stage PCR assays for gltA and ompA were the only ones which allowed the amplification of rickettsiae from blood samples and biopsies. In accordance with the European Guidelines,18 our data showed that ticks attached to humans, skin biopsy and EDTA-treated blood specimens are valuable clinical samples for Rickettsia speciation.
Regarding identification of Rickettsia spp., our results showed that ompB PCR assays are the less species-specific methods. In our environment and according to our experience, ompA PCR is the most valuable tool for the identification of Rickettsia species in clinical samples, taking into account the clinical syndrome of the patient.
This study shows that to investigate which Rickettsia species is implicated in the clinical syndrome may be a difficult problem to solve. For example, if we had only chosen the htrA PCR, we could have misdiagnosed Rocky Mountain spotted fever in Europe. The same lack of accuracy for species identification was found testing samples with other assays such as the16S rDNA PCR. As it has been confirmed with our results, 16S DNA and htrA genes are genus level markers for rickettsiae and they are not informative genes for speciation. According to Raoult et al. at least two different genes must be used to implicate a new Rickettsia sp. in human pathology.29 We have used multi-locus sequence typing for naming or involving a new Rickettsia sp. as human pathogen.9,14,34 This makes the use of real-time PCR assays for a specific diagnosis of rickettsioses difficult, although it may be a good strategy to increase the sensitivity.
In summary, the PCR-based amplification method is a useful rickettsial diagnostic tool in the early phase of the illness. Rickettsiae can be detected from clinical samples including skin biopsies, EDTA-blood and sera specimens by PCR assays. The three sequential PCR assays here investigated (ompB, gltA and ompA) represent useful tools for the molecular diagnosis of human rickettsioses. As diagnostic algorithm for rickettsiosis, we recommend performing gltA and ompB PCR assays, followed in positive samples by ompA PCR and nucleotide sequence analysis. ompB PCR is an effective method for the first screening since it allows detecting a high percentage of positive samples. The ompA PCR assay is the most accurate method to diagnose and to implicate a new Rickettsia species in human pathology. Nevertheless, epidemiology, clinical symptoms and the vector implicated in the infection have to be taken into account for the final diagnosis of rickettsioses.
FundingThis work was supported in part by a grant from “Instituto de Salud Carlos III” (EMER 07/033), Ministerio de Ciencia e Innovación (Spain).
Conflict of interestThe authors have no conflict of interest to declare.
We thank Fátima Bacellar, Rita de Sousa and Natacha Milhano from Instituto Nacional de Saúde Dr Ricardo Jorge (Portugal), for supplying Rickettsia-positive controls.