During a community methicillin-resistant Staphylococcus aureus (MRSA) nasal colonization study, an MRSA strain with vancomycin hetero-resistance (h-VISA) was isolated from a five year-old girl with tetralogy of Fallot without previous exposure to vancomycin. An extended nasal colonization study was performed on all her close relatives.
ResultsOnly the patient and her sister were colonized by an h-VISA MRSA strain (clone USA 700, ST72, t148, agr 1 and SCCmec IVa). Mupirocin decolonisation was effective in the elder sister. A new nasal decolonisation in the younger girl using fusidic acid was also successful. However, after decolonisation both sisters were colonized by a methicillin-susceptible S. aureus (ST30, t012 and agr 3) previously isolated from their mother's nostrils.
ConclusionAs S. aureus have a great capacity to spread among people in close contact, knowledge of a patients’ colonization status, tracing contacts, and a correct management are critical issues for the successful containment of multiresistant staphylococci.
Durante un estudio comunitario de colonización nasal, hemos aislado Staphylococcus aureus resistente a la meticilina (SARM) con heterorresistencia a vancomicina (hVISA) en una niña de 5 años, que padecía una tetralogía de Fallot, que no había sido tratada previamente con vancomicina.
ResultadosEste hallazgo nos llevó a extender el estudio de colonización a sus familiares más cercanos. De estos, solo su hermana mayor fue colonizada por esta cepa SARM hVISA (clon USA 700, ST72, t148, agr 1 y SCCmecIVa). La descolonización con mupirocina fue eficaz en el caso de la hermana, pero un tratamiento con ácido fusídico fue necesario para eliminar la colonización nasal de la paciente. Sin embargo, después de la descolonización, ambas hermanas fueron colonizadas por una cepa de S. aureus sensible a meticilina (ST30, t012 y agr 3), que previamente había sido aislada de las fosas nasales de su madre.
ConclusiónS. aureus tiene una gran capacidad de diseminación entre personas en estrecho contacto, por lo que el conocimiento del estado de colonización de los pacientes, la evaluación de la colonización nasal de los contactos y una aproximación terapéutica correcta son esenciales para la contención de la diseminación de cepas de estafilococos multirresistentes.
Staphylococcus aureus is a Gram-positive coccus that frequently colonizes human skin and mucosa, and it is associated to many life-threatening infections. Besides, this microorganism produces numerous virulence factors, toxins such as Panton-Valentine leukocidin (LPV) and the toxic shock syndrome toxin 1 (TSST-1), and it has a great ability to evolve rapidly acquiring resistance to current antimicrobial drugs which confers an additional capacity for survival. Methicillin-resistant S. aureus (MRSA) is prevalent in health care associated infections and it also can be a relevant aetiological agent in community infections. Vancomycin has been the drug of choice to treat infections caused by MRSA strains until 1996 when a community acquired pneumonia case due to a vancomycin intermediate S. aureus (VISA) was reported in Japan.1 This strain had a thicker cell wall, produced a larger amount of penicillin binding protein (PBP) 2 and 2a, and showed a vancomycin minimum inhibitory concentration (MIC) of 4μg/ml. However, this MRSA strain did not present vanA, vanB or vanC genes that confer high-level resistance to vancomycin in Enterococcus.2 Later on, some strains have been referred as heteroresistant vancomycin-intermediate S. aureus (h-VISA) for including subpopulations with vancomycin MICs between 1 and 2μg/ml mixed with other VISA subpopulations. Strains with vancomycin MICs≥2μg/ml have a decreased susceptibility and have been associated with therapeutic failures, despite the use of high doses of vancomycin.3 The emergence of vancomycin resistant S. aureus (VRSA) is mostly associated with the acquisition of vanA gen, which probably transferred by conjugation from a vancomycin-resistant Enterococcus strain. However, VISA and h-VISA strains are not linked with these genes and it is probably related to complex genetic and cell wall changes.4–6 However, Hiramatsu et al.,7 consider that h-VISA is a precursor of VISA after a selective pressure by the use of betalactams and glycopeptides. h-VISA and VISA reports have increased in Europe, Asia and USA since 1996 and most of them are MRSA.8–10
Patients at greatest risk include those who have previously been treated with glycopeptides, or underwent to cardiovascular surgery, non-vascular prosthesis implantation surgery and dialysis.11,12 Treatment for these strains with reduced susceptibility to vancomycin includes alternatives such as linezolid, daptomycin, and trimethoprim/sulfamethoxazole.13
The aim of this study was to report a case of intra–familiar transmission of an unusual h-VISA MRSA clone, colonizing two sisters without previous exposition to vancomycin.
Patients, materials and methodsPatientsDuring a community based colonization study, conducted between 2010 and 2012 in the villages of Plentzia, Gorliz and Barrika (Bizkaia, Basque Country, North of Spain), a MRSA strain was isolated from the anterior nares of a five-year-old Spanish girl suffering from tetralogy of Fallot and cardiovascular surgery. No other co-morbidities were reported in her clinical history. The girl received oral amoxicillin and intravenous cefuroxime in the past. To confirm these results and following the existing infection control program, new samples were obtained from the child and her close relatives (father, mother, grandparents and a seven years old sister): Twenty-five samples were studied from both anterior nares of the two sisters and their close relatives.
Sample collection and Staphylococcus aureus isolation and identificationNasal samples were collected in Amies Portagerm transport (BioMérieux, France) and inoculated as soon as they arrived at the laboratory onto salt-mannitol agar plates (BBL, France). After 48h incubation at 36±1°C, all the strains with different morphology were subcultured onto chromogenic media – chromID S. aureus (BioMérieux) and CHROMagar Staph aureus (CHROMagar, France) – and incubated for another 48h at 36±1°C. All presumptive S. aureus strains were plated onto blood agar (BioMérieux) and subsequently identified using Gram staining, catalase, tube coagulase (Staph-ase, BioMérieux) and latex agglutination (Slidex Staph Plus, BioMérieux) tests, and the ID 32 Staph biochemical gallery (BioMérieux).
MRSA detection and antimicrobial susceptibility testingMethicillin resistance was studied using chromogenic medium CHROMagar MRSA (CHROMagar), a latex agglutination test (Slidex MRSA, BioMérieux), and cefoxitin disks according to the Clinical Laboratory and Standards Institute guidelines (CLSI). Moreover, the presence of mecA gene was detected by using conventional polymerase chain reaction (PCR).14 Disk diffusion and broth microdilution antimicrobial susceptibility testing of S. aureus isolates was performed following the M02 and M07 CLSI guidelines.15,16 For testing high and low level resistance to mupirocin, two disks of 5 and 200μg (Oxoid, UK) were used.15 Gram-positive CLSI Microscan® panel (Siemens Healthcare, Germany) was used for the broth microdilution testing.16 In addition, to identify h-VISA, VISA or VRSA strains, teicoplanin and vancomycin MICs were also determined by a macro E-test procedure (BioMérieux).
Toxin productionPresence of luk-PV and tsst gene that encode the PVL and the TSST-1 toxins, respectively, were studied by PCR17,18 using ATCC 49775 and NCTC 7428 strains as positive controls for each technique.
Molecular typingMolecular typing of all S. aureus isolates was performed using several techniques which included Pulsed Field Gel Electrophoresis (PFGE), Multilocus Sequence Typing (MLST), staphylococcal protein A typing (spa typing), accessory gene regulator (agr) typing and staphylococcal cassette chromosome mec (SCCmec) typing.19–23
Patient managementAll patients colonized with MRSA were treated topically with mupirocin calcium cream in their anterior nares.
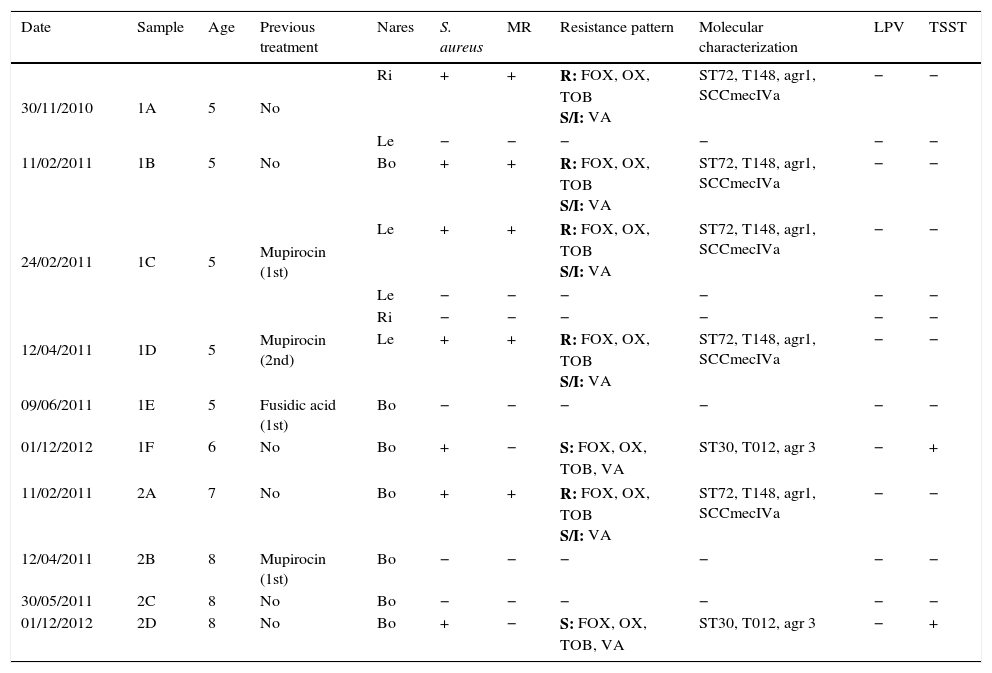
ResultsTable 1 summarizes the staphylococcal colonization in both sisters’ nares throughout the study period and the phenotypic and genotypic characteristics of the clinical isolates. The five-year-old girl, her elder sister, their mother and maternal grandfather were colonized by S. aureus in both nares. Conversely, the father, the paternal grandparents and the maternal grandmother were not colonized by S. aureus. Both sisters were colonized by MRSA strains, whereas their mother and maternal grandfather were colonized by methicillin-susceptible S. aureus (MSSA) strains.
Staphylococcal colonization in both sisters’ nares throughout the study period.
| Date | Sample | Age | Previous treatment | Nares | S. aureus | MR | Resistance pattern | Molecular characterization | LPV | TSST |
|---|---|---|---|---|---|---|---|---|---|---|
| 30/11/2010 | 1A | 5 | No | Ri | + | + | R: FOX, OX, TOB S/I: VA | ST72, T148, agr1, SCCmecIVa | − | − |
| Le | − | − | − | − | − | − | ||||
| 11/02/2011 | 1B | 5 | No | Bo | + | + | R: FOX, OX, TOB S/I: VA | ST72, T148, agr1, SCCmecIVa | − | − |
| 24/02/2011 | 1C | 5 | Mupirocin (1st) | Le | + | + | R: FOX, OX, TOB S/I: VA | ST72, T148, agr1, SCCmecIVa | − | − |
| Le | − | − | − | − | − | − | ||||
| 12/04/2011 | 1D | 5 | Mupirocin (2nd) | Ri | − | − | − | − | − | − |
| Le | + | + | R: FOX, OX, TOB S/I: VA | ST72, T148, agr1, SCCmecIVa | − | − | ||||
| 09/06/2011 | 1E | 5 | Fusidic acid (1st) | Bo | − | − | − | − | − | − |
| 01/12/2012 | 1F | 6 | No | Bo | + | − | S: FOX, OX, TOB, VA | ST30, T012, agr 3 | − | + |
| 11/02/2011 | 2A | 7 | No | Bo | + | + | R: FOX, OX, TOB S/I: VA | ST72, T148, agr1, SCCmecIVa | − | − |
| 12/04/2011 | 2B | 8 | Mupirocin (1st) | Bo | − | − | − | − | − | − |
| 30/05/2011 | 2C | 8 | No | Bo | − | − | − | − | − | − |
| 01/12/2012 | 2D | 8 | No | Bo | + | − | S: FOX, OX, TOB, VA | ST30, T012, agr 3 | − | + |
Bo: both nares; Le: left; Ri: right. MR: methicillin resistant; R: resistant; S: susceptible; S/I: h-VISA strain. Resistance patterns: All strains were resistant in vitro to penicillin and susceptible to clindamycin, erythromycin, gentamicin, levofloxacin, linezolid, mupirocin, ofloxacin, quinupristin-dalfopristin, rifampicin, sulfamethoxazole, teicoplanin, and tetracycline. FOX: cefoxitin; OX: oxacillin; TOB: tobramycin; VA: vancomycin.
MRSA strains showed a 14mm inhibition zone diameter around the vancomycin disk and E-test MIC values for vancomycin and teicoplanin equal to 2μg/ml. However, the microdilution panel showed a vancomycin MIC of 4μg/ml and a teicoplanin MIC equal to 2μg/ml. Broth dilution and Etest macromethod vancomycin MICs from different colonies of both girls MRSA strains confirmed that they were h-VISA. In addition, MRSA and MSSA strains were resistant to tobramycin except for those MSSA strains yielded from the sisters’ control samples after mupirocin decolonization that were only resistant to penicillin G. None of the S. aureus isolates was LPV positive. However, the last MSSA strains isolated from both sisters, the mother and the maternal grandfather contained the tsst gene.
MRSA strains isolated from both sisters belonged to the same clone and showed identical PFGE profile: USA 700 like, ST72, t148, agr 1 and SCCmec IVa. However, there were slightly differences among the mother's MSSA strain type (ST30, t012 and agr 3) and the maternal grandfather's MSSA strain, having the same ST and agr type (ST30 and agr 3), but different spa type (t1515 instead of t012). Regarding the MSSA strains isolated from both sisters, the one isolated from the five-year-old girl belonged to the same clone as her mother's MSSA strain (ST30, t012 and agr 3) but the MSSA strain isolated from the seven years old girl was ST 34, t089 and agr 3. Therefore, pulsotypes of the MSSA strains were similar but not the same: MSSA strains isolated from the mother and the youngest girl were genetically related to the MSSA strain isolated from the grandfather. However, the MSSA strain isolated from the eldest girl after mupirocin decolonization therapy was genetically different.
Both girls were treated with mupirocin calcium cream three times a day during five days according to the established infection control protocol. A week after therapy completion, new nasal samples were obtained and a MRSA strain identical to those described before was isolated from the right nare of the youngest girl. After a second unsatisfactory decolonizing treatment with mupirocin, a new decolonization treatment was proposed using fusidic acid with successful response and no isolation of MRSA or MSSA strains. Several months later, new control samples were obtained from both sisters and showed that both of them were re-colonized by different MSSA strains.
DiscussionIn the current study, we described the transmission of a vancomycin heteroresistant community associated MRSA strain that colonized the anterior nares of two Spanish girls. This h-VISA and MRSA strain was isolated in several samples obtained from both sisters, suggesting a transmission from one to the other. Due to the previous clinical history of the youngest girl, probably she was colonized during one of her hospitalizations and afterwards she transmitted the strain to her sister by direct contact. Besides, this study also emphasizes the importance of making a good infection control interview and of a tracing study when an unusual S. aureus strain is detected and highlights other transmissions such as the MSSA strain previously isolated from the mother and afterwards isolated from her five years old daughter apart from complex relationships among different sort of strains. The molecular typing made during the contact tracing performed in this study allowed us to understand the genetic similarities or differences between all MSSA and MRSA strains, and help us in constructing different hypothesis of transmission and a possible common or different origin of the strains. The great capacity of S. aureus to be transmitted locally among people who are close in contact has been claimed.24,25 Host characteristics are also important to be colonized.26 When a niche is occupied by one bacterium, another bacterium is not capable of replacing it without any help and a successful replacement requires the reduction or even elimination of the primary strain.27,28 In the current study, both girls were re-colonized by two different MSSA strains, after a successful MRSA decolonization.
Several studies pointed that vancomycin heteroresistance is increasing but still rare.12,29 Furthermore, this resistance occurs mainly in MRSA strains, hospitals and patients previously treated with vancomycin, and/or having different risk factors such as cardiovascular surgery and dialysis. The current study h-VISA strain was isolated from the anterior nares of a young girl never treated with glycopeptides. Her only remarkable risk factor was a cardiac surgery to correct a tetralogy of Fallot. Therefore, this strain had similar characteristics (young patients, no presence of vanA, vanB or vanC genes, previous cardiac surgery and hospital staying) to those h-VISA strains described previously.6,12 Hiramatsu et al. indicated that either previous treatment with betalactams or glycopeptides could develop a vancomycin heteroresistance2,30 and suggested that this vancomycin heteroresistance could be linked to the conversion mechanisms of methicillin resistance.31 Although Park et al. described that the Korean ST72-SCCmecIV strains isolated from adult patients with MRSA bacteraemia were more prone to show decreased vancomycin susceptibility, there are several differences with our study. Their strains were isolated from adult patients and not from paediatric ones like ours. A second remarkable difference is that all the strains typed in the mentioned study were not colonizing the patient rather than causing him a potential life-threatening infection.32 Children nasal colonization is a key point in our study; as the same MRSA strain but without reduced susceptibility to vancomycin has been detected colonizing adult people in Basque Country. Besides, local and personal epidemiological factors may play a role in the acquisition and expression of virulence factors and antimicrobial resistance mechanisms by genetically related S. aureus strains. Therefore, the young girl probably acquired this h-VISA strain during one of her hospital stays where she received multiple treatments with betalactams although she was never treated with vancomycin. Besides, h-VISA strain was transmitted to her eldest sister who neither was at the hospital nor received antimicrobial treatments during the last three years.
MRSA isolates were classified as clone ST72, t148, agr 1 and SCCmec type IV, and had a PFGE pattern related with USA 700. This ST72 MRSA clone was first described in South Korea,33 being rare elsewhere in the world. ST72 has not been reported in hospital or community based studies performed the Basque Country. In neighbour regions of Portugal and Spain, some of the MRSA strains isolated from patients between 2002 and 2009 belonged to ST72 – t148 clade, but none of these isolates were h-VISA.34,35 A recent study published by Potel et al. highlights that this clade is infrequent in other countries except South Korea.36 However these authors found an increasing prevalence of this clon in S. aureus isolated from community infections and a displacement effect in the local molecular epidemiology of S. aureus infections not related to demographic conditions. Although these authors discussed the potential role of commercially imported meat in the introduction of new MRSA clones (like the ST72, t148, agr 1 and SCCmec type IV) to explain such an observation, the limited surveillance data in this field prevent these authors from getting a real evidence to confirm such a hypothesis.36 Moreover, the MSSA ST30 – t012 clone isolated in our study is genetically related to the Southwest Pacific clone that has been the most prevalent one among community and hospital settings in Portugal between 1992 and 2011.35 This MSSA clade has also been widely isolated in England, Germany, Holland and Spain.37–42
Unsuccessful treatment in the five years old girl may indicate that treatment was not appropriate or that the amount of mupirocin was inadequate. Moreover, patients can be persistently colonized with MRSA in a special deeper niche that hinders the antimicrobial action. Despite which of the mentioned statements is true, nasal decolonization mupirocin treatment eradicated the h-VISA MRSA strain in the seven years old sister. This evidence could suggest that the colonization in the eldest sister was transient. Salgado et al. isolated some strains with low mupirocin resistance after treatment with this drug.42 In the current case, we did not find low or high level resistance to mupirocin.
MRSA nasal decolonization procedure using mupirocin is controversial.41–44 Wertheim et al.,42 indicate that there is little evidence that nasal decolonization is effective except in patients on dialysis or surgery. Furthermore, these patients are often re-colonized in a short period of time.41 However, a recent randomized study conducted in several adult ICUs in USA concluded that universal decolonization was more effective that other studied interventions in reducing the rates of MRSA clinical isolates and nosocomial bloodstream infections.45 In our case, MRSA nasal decolonization therapy was initially effective but both sisters were afterwards colonized by a different MSSA strain remarking the previously described findings. However, decolonization in the current case, where the strain was a highly resistant one, was mandatory in order to prevent the transmission to other people. In our study, the h-VISA MRSA strain did not have the tsst and lukPV genes, important biomarkers of community-acquired MRSA (CA-MRSA strains), although Dinges et al.,46 reported that many CA-MRSA strains presented lukPV but no tsst.
In conclusion, h-VISA and MRSA strains can be easily transmitted by personal contact, raising the need to develop appropriate infection control measures to prevent its spread. Not only healthcare personnel but also visitors and patients who access a health centre should be aware of the importance of infection control measures to minimize the transmission of these strains.
Transparency declarationsIn the past 5 years, E.E. has received grant support from Astellas Pharma, and Pfizer. G.Q. has received grant support from Astellas Pharma, Gilead Sciences, Pfizer, and Merck Sharp and Dohme. He has been an advisor/consultant to Merck Sharp and Dohme, and has been paid for talks on behalf of Abbvie, Astellas Pharma, Gilead Sciences, Merck Sharp and Dohme, and Pfizer SLU.
FundingThis work was supported by Consejería de Educación, Universidades e Investigación (GIC12 210-IT-696-13) and UPV/EHU (UFI 11/25).
Conflict of interestThe authors declare no conflict of interest.