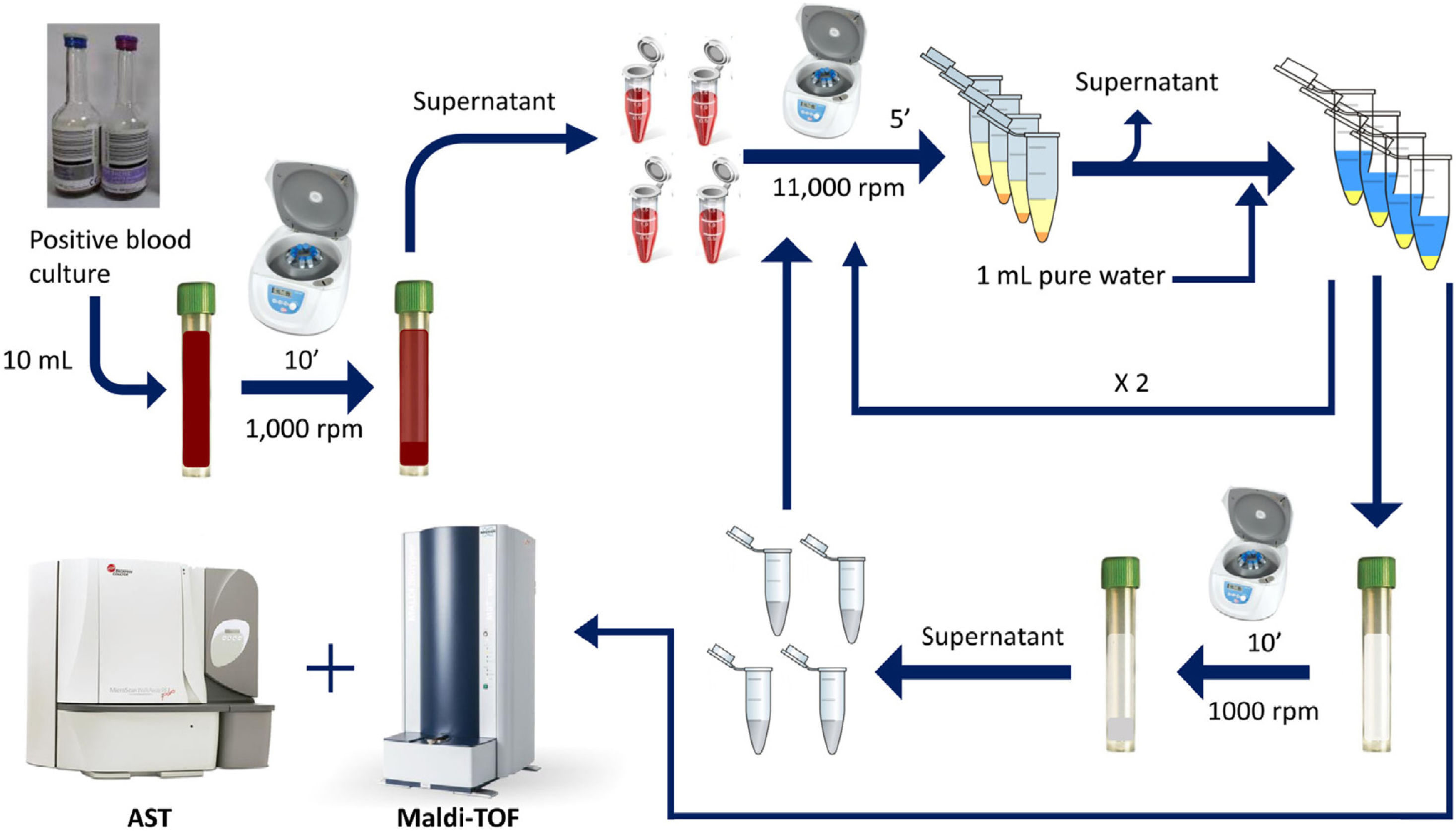
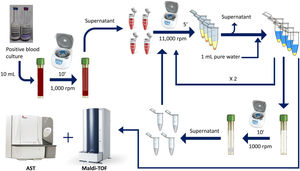
This study proposes a simple and rapid method for both bacterial identification and direct antimicrobial susceptibility testing (AST) by using MALDI-TOF and a double differential centrifugation-wash procedure from positive blood cultures.
MethodsFifty-two positive blood cultures (37 gramnegative bacilli and 15 grampositive cocci) were studied by two methods for identification and AST: a reference method, and the rapid MALDI-TOF method obtaining a purified pellet by using a double differential centrifugation procedure.
ResultsA total of 1101 MIC values (mg/l) were interpreted according to EUCAST clinical breakpoints and compared using the two methods simultaneously. Discrepancies in 81 MIC values (7.35%) were detected. By analyzing standard parameters, we obtained 98.28% essential agreement and 92.65% categorical agreement considering all isolates tested.
ConclusionThis method provides rapid bacterial identification and AST, offering definitive results 24–48h earlier than the conventional method (p<0.001) and improving the turnaround time in blood culture diagnostics, especially in laboratories without 24-h on-call.
Este trabajo propone un método sencillo, rápido y barato de identificación bacteriana y sensibilidad antibiótica utilizando MALDI-TOF y una doble centrifugación diferencial a partir de hemocultivo positivo.
MétodosSe estudiaron 52 hemocultivos positivos (37 bacilos gramnegativos y 15 cocos grampositivos). Se compararon 2 métodos: un método convencional de identificación y determinación de sensibilidad a antibióticos automatizada partiendo de colonia crecida, y un método rápido utilizando MALDI-TOF, caracterizado por la obtención de un pellet purificado procedente de un hemocultivo positivo, mediante un procedimiento basado en una doble centrifugación diferencial.
ResultadosSe analizaron y compararon 1.101 valores de CMI (mg/l) de acuerdo con los criterios establecidos por EUCAST y obtenidos por ambos métodos. Se detectaron discrepancias en 81 valores de CMI (7,35%). Considerando todos los aislados, la concordancia esencial fue del 98,28% y la concordancia categórica del 92,65%.
ConclusiónEste método proporciona resultados de identificación y sensibilidad a antibióticos definitivos 24-48h antes que un método convencional (p<0,001), mejorando el tiempo de respuesta en el diagnóstico microbiológico de bacteriemias, especialmente en laboratorios sin servicio de guardias de 24h.
Clinical microbiology laboratories try to get accurate and timely antimicrobial susceptibility testing (AST) data in order to achieve an adequate treatment. A rapid and early determination of adequate antibiotic susceptibility results is crucial for management of patients with bacteremia, particularly important in patients with sepsis.1,2 To select proper antibiotic regimens, it is necessary to correctly identify the etiological agent from the bloodstream infection, as well as to determine AST as soon as possible. A reduction in the turnaround time for microbial identification and AST to less than one working day reduces morbidity and mortality, as well as the overall costs for healthcare systems.3 Moreover, anticipating AST results to the clinicians has a significant impact on patient therapy and could be implemented as a part of antimicrobial stewardship programs.4 Several studies have recently focused on the development of methods to identify bacteria and to get AST results directly from positive blood cultures earlier than those obtained with standard methodologies.5,6 Some of them agree that the lysis/centrifugation method provides the most accurate results.7 In this study, we aimed to develop a simple and inexpensive blood sample preparation method for both, bacterial identification and direct AST of microorganisms in patients with bacteremia. This is based on a double differential centrifugation-wash procedure to get a purified pellet from a positive blood culture, without needing the addition of lysis buffer that could interfere on bacterial viability. The results were compared with those obtained through conventional laboratory culture-dependent sample preparation procedures.
Material and methodsStudy designThe study was conducted at Getafe University Hospital (Madrid, Spain) from April 2019 to September 2019. Randomly selected positive blood cultures were processed by two different methods: a conventional identification by MALDI-TOF and automated microdilution AST method, considered as the reference by using bacterial colonies grown in solid media (see below), and a rapid method obtaining a purified pellet. Aerobic and anaerobic blood culture bottles from patients with suspected bloodstream infection were incubated in a BACTEC™ FX instrument (Becton & Dickinson, Erembodegem, Belgium), according to the manufacturer's instructions. Aerobic bottles were not prioritized over anaerobic ones for processing.
Bacterial strainsThe microorganisms analyzed in this study were obtained from anonymized isolates of blood samples. Fifty-two positive blood cultures were randomly selected for processing: 30 Enterobacterales (19 Escherichia coli (5 ESBL-producers), 3 Klebsiella pneumoniae, 1 Klebsiella variicola, 2 non-typhi Salmonella, 3 Klebsiella aerogenes, 1 Serratia marcescens, 1 Enterobacter cloacae), 5 non-fermenters gramnegative bacilli (4 Pseudomonas aeruginosa, 1 Stenotrophomonas maltophilia), 6 Staphylococcus spp. (5 Staphylococcus aureus, 1 Staphylococcushaemolyticus), 4 Enterococcus spp. (2 Enterococcus faecium, 2 Enterococcus faecalis), 5 Streptococcus spp. (3 Streptococcus gallolyticus, 1 Streptococcus gordonii, 1 Streptococcus constellatus) and 2 anaerobic gramnegative bacilli (2 Bacteroides fragilis).
Reference methodPositive blood culture bottles were directly processed for Gram stain and subcultured on solid media (blood agar, chocolate agar, MacConkey agar and anaerobic agar). All plates were incubated at 35°C with 5% CO2 and, after overnight incubation, the isolated colonies were subjected to conventional laboratory identification by MALDI-TOF MS and AST using MicroScan Walkaway microdilution panels: NM EN-52 MIC Panel for Enterobacterales and non-fermenters GNB, PM33 MIC Panel for staphylococci/enterococci, MSTRP+6 MIC Panel for streptococci (Beckman Coulter International S.A., Nyon Switzerland) and gradient concentration strips (MIC Test Strip, Liofilchem S.r.l., Italy) for anaerobic GNB following the manufacturer's guidelines. If the MALDI-TOF score of identification was higher than 1.7 the identification of the isolate was accepted.
Rapid method by double differential centrifugationThe same positive blood cultures bottles described above were simultaneously processed using a double differential centrifugation wash-procedure based in low-speed/high-speed method (see Fig. 1 and legend). In order to inoculate the MicroScan panels, the purified pellet was allowed to dry at room temperature for 5min and was punched by using the PromptTM Inoculation System Wands, removing the standardizer prior to the preparation of the final bacterial suspension. The inoculum of viable bacteria was tested by sub-culturing serial dilutions of a 100μl suspension onto a blood agar plate, which was incubated 24h at 37°C. Bacterial inoculum was stablished as ≥105cfu/ml in all the isolates.
Double differential centrifugation wash-procedure to obtain the purified pellet. A 10-ml sample of a positive blood culture was first centrifuged at 1,000rpm for 10min, followed by transferring 6ml of the supernatant into four 1.5ml conical-bottomed tubes. The transferred supernatants were centrifuged at 11,000rpm for 5min, and washed twice with purified water. The resulting bacterial pellet was resuspended into 10ml of purified water and was again centrifuged at 1000rpm (second differential centrifugation) for 10min, followed by transferring again into four 1.5ml conical tubes. The transferred supernatants were finally centrifuged at 11,000rpm for 5min and the resulting pellet was subjected to MALDI-TOF identification and direct AST.
A total of 1101 MIC values (mg/L) interpreted according EUCAST clinical breakpoints,8 were analyzed and compared using the standard parameters: categorical agreement (MICs matched, CA), essential agreement (MICs not matched but same interpretation by EUCAST, EA), very major errors (false susceptibility, VME), major errors (false resistance, ME) and minor errors (intermediate versus susceptible or resistant, mE)). Error levels were expressed as percentages and calculated considering the conventional method as the reference. The procedure was validated according to Clinical Microbiology Procedures in which at least 90% agreement in AST results is mandatory.9 Values for the kappa coefficient were interpreted according to the Landis and Koch classification.10
ResultsAll the pellets purified from blood cultures were identified with a score higher than 2 and there were no discrepancies in genus or species results in relation with the conventional method. A total of 1101 MIC values were obtained and discrepancies were detected in 81 MIC (7.35%) in relation to the reference method. By analyzing standard parameters, we detected 98.28% EA and 92.65% CA considering all isolates tested.
In the Enterobacterales group, there were discrepancies between both methods in 34 out of 813 MIC results (4.18%), 0.36% being VME, 0.24% being ME and 1.47% being mE (Table 1A). The EA and CA for all the antibiotics tested were 97.91% and 95.82%, respectively. In the non-fermenters group, there were discrepancies in 18 out of 57 MIC results (31.57%) but no VME, ME or mE were detected and, therefore, the final interpretation was not altered. The EA and CA for all the antibiotics tested were 100% and 68.43%, respectively.
Comparison of the discrepant results obtained between the reference method and the rapid method for Enterobacterales (1A) and Enterococcus spp. (1B).
| Antibiotic | Reference method (number of isolates) | Rapid method (number of isolates) | Kappa | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EUCAST interpretation MIC | Changes in MIC according to EUCAST interpretation | |||||||||||
| S | I | R | Stay S | Stay R | VME | ME | mE | |||||
| R → S | S → R | R → I | I → S | I → R | S → I | |||||||
| 1A | ||||||||||||
| Enterobacterales (N=30) | ||||||||||||
| Amoxicillin/clavulanic acid | 21 | 0 | 9 | 20 | 8 | 1 | 1 | – | – | – | – | 0.841 |
| Ticarcillin | 10 | 0 | 20 | 10 | 19 | – | – | 1 | – | – | – | 0.929 |
| Piperacillin | 13 | 1 | 16 | 13 | 15 | 1 | – | – | 1 | – | – | 0.871 |
| Piperacillin/tazobactam | 25 | 2 | 3 | 25 | 3 | – | – | – | – | 2 | – | 0.769 |
| Ceftazidime | 23 | 1 | 6 | 22 | 6 | – | – | – | – | – | 1 | 0.903 |
| Cefepime | 25 | 1 | 4 | 24 | 3 | – | – | 1 | 1 | – | 1 | 0.655 |
| Aztreonam | 24 | 0 | 6 | 23 | 6 | – | – | – | – | – | 1 | 0.904 |
| Ertapenem | 29 | 1 | 0 | 29 | 0 | – | – | – | 1 | – | – | 0.967 |
| Gentamicin | 27 | 0 | 3 | 26 | 3 | – | – | – | – | – | 1 | 0.841 |
| Tobramycin | 24 | 0 | 6 | 23 | 6 | – | – | – | – | – | 1 | 0.904 |
| Nitrofurantoinb | 19 | 0 | 0 | 18 | 0 | – | 1 | – | – | – | – | 0.947 |
| Mecillinama | 17 | 0 | 6 | 17 | 5 | 1 | – | – | – | – | – | 0.881 |
| 1B | ||||||||||||
| Enterococcus spp. (N=4) | ||||||||||||
| Ciprofloxacin | 1 | 0 | 3 | 1 | 2 | – | – | 1 | – | – | – | 0.556 |
| Levofloxacin | 1 | 0 | 3 | 1 | 2 | 1 | – | – | – | – | – | 0.500 |
Concerning grampositive cocci (GPC), there were discrepancies in 29 out of 219 MIC results (13.24%), 0.45% being VME and 0.45% being mE (Table 1B). No ME were detected. The EA and CA for antibiotics tested in GPC were 99.09% and 86.76%, respectively. MIC results with no VME, ME and mE are summarized in the tables of supplementary material (supplementary Tables 1 and 2).
In anaerobic GNB (2 B. fragilis) no discrepancies were observed in gradient concentration strips results (data not shown).
Taken together, this AST method was in agreement with the standard AST method (for <100 isolates tested, <5% VME and ME and <10% combined ME and mE) and meets the required criteria to be validated, considering all isolates tested.9 This method offers definitive results 24–48h earlier than the conventional method (p<0.001).10
DiscussionThe proposed protocol offers several advantages over other commercial techniques needing more complex equipment that not always are available in all laboratories.11,12 The technique described here is based on a simple double differential centrifugation-wash procedure used to obtain a clean purified pellet avoiding the impact of a lysis buffer on the viability of bacteria.
Previous studies have reported slightly higher sensitivity in identification and AST results in bacterial pellets from lysis/centrifugation over pellets obtained by differential centrifugation.7,13 At this point, it is important to take into account that a second differential centrifugation can solve this problem, especially important in pellets obtained from anaerobic blood culture bottles in which many blood components are not adequately eliminated after a simple differential centrifugation (personal observation). The results obtained in this study show no errors in identification when compared with the reference method from bacterial colonies, similar to those described in previous studies.5,14 In addition, what is even more important is that we describe a method that provides definitive laboratory results at 24h, and 48h in the case of anaerobic bacteria, earlier than when using the conventional method, improving the turnaround time in the diagnosis of bacteremia. This is especially important in the case of sepsis, as inadequate antimicrobial treatment is associated with unfavorable outcomes and an early and adequate treatment is essential.15 The method described in our study, has a notable advantage, especially in laboratories that do not have 24-h on-calls, since definitive MIC results can be available within 18h after detecting a positive blood culture at the end of the previous day, without the need to have the bacterial colony grown on the culture plate.
Two VME were detected in an ESBL-producing E. coli isolate related to amoxicillin/clavulanate and mecillinam MICs. Also, a VME was detected in a K. aerogenes isolate related to piperacillin MIC. It is important to emphasize that, from the clinical point of view, both mecillinam and piperacillin are not used as antibiotic treatments for infections caused by these bacteria. Two ME were also detected in E. coli isolates related to amoxicillin/clavulanate and nitrofurantoin MICs. Only a VME was detected in an E. faecium isolate related to levofloxacin MIC. Several studies have reported similar results.5,6
The study has several limitations; the relatively small number of isolates analyzed, especially non-fermenters and anaerobic gramnegative bacilli. Although no errors were detected in P. aeruginosa and B. fragilis isolates, more isolates should be analyzed to conclude that this tool is effective with these microorganisms to get reliable results. Another important limitation is that only 26% of E. coli isolates were ESBL-producers (5 out 19 E. coli isolates) and no ESBL-producers were analyzed in the remaining Enterobacterales (different from E. coli). Thus, it would be desirable to analyze more isolates with this and other phenotypes, such as plasmidic AmpC and carbapenemase-producers. It is also important to clarify that this method is not a valuable tool to identify and to get correct AST results in polymicrobial blood cultures as results might be erratic.
To conclude, this method provides rapid bacterial identification and AST data that enable the clinicians to choose an appropriate antibiotic treatment, offering definitive laboratory results 24–48h earlier than when using the conventional method and improving the turnaround time in the diagnosis of bacteremia, especially useful in laboratories without an established 24-h on-call service.
Author contributionArana DM was responsible for the organization and coordination of the trial and also was the chief investigator and responsible for the data analysis. Arana DM and Hernández Y developed the trial design. All authors contributed to the writing of the final manuscript.
Ethical approvalNot required.
FundingThis research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Conflict of interestsNone declared.