The increase in sexually transmitted infections (STI) caused by Neisseria gonorrhoeae (NG) worldwide, together with the decrease in antibiotic susceptibility, forced us to understand the epidemiology of gonococcal infection.
MethodsThe GONOvig project analyzed, comparatively following CLSI and EUCAST criteria, the antibiotic susceptibility of 227 NG strains collected in thirteen representative hospitals of the Valencia Community (CV) between 2013 and 2018. Additionally, molecular typing of 175 strains using the NG multi-antigen sequence typing technique (NG-MAST) was performed.
ResultsHigh rates of resistance to tetracycline (38.2% by CLSI and 50.9% by EUCAST) and ciprofloxacin (49.1% CLSI and 54% EUCAST), and low percentages of resistance to spectinomycin (0%), cefixime (0.5% CLSI but 5.9% EUCAST), and ceftriaxone (1.5% CLSI and 2.4% EUCAST) were detected. Azithromycin resistance was 6% (both CLSI and EUCAST). Molecular analysis revealed the presence of 86 different sequence types (ST), highlighting ST2992 (7.4%), ST3378 (6.9%), ST2400 (4.6%) and ST13288 (6.9%), which was associated with resistance to cefixime (P=.031). The main genogroups (G) were G1407 (13.1%), G2992 (10.3%), G2400 (6.3%) and G387 (3.4%). G1407 and G2400 were associated with resistance to ciprofloxacin (P<.03).
ConclusionLow resistance to ceftriaxone, a worrying resistance to azithromycin and a wide variety of circulating sequence types have been detected, some of which show correlation with certain resistance profiles.
El aumento de las infecciones de transmisión sexual producidas por Neisseria gonorrhoeae (NG) a nivel mundial, junto con la disminución de la susceptibilidad antibiótica, obliga a profundizar en la epidemiologia de la infección gonocócica.
MétodosEl proyecto GONOvig analizó, comparativamente siguiendo criterios CLSI y EUCAST, la sensibilidad antibiótica de 227 cepas de NG recogidas en trece hospitales representativos de la Comunidad Valencia (CV) entre los años 2013 y 2018. Adicionalmente, se pudo realizar la tipificación molecular de 175 cepas mediante la técnica NG multi-antigen sequence typing (NG-MAST).
ResultadosSe detectaron elevadas tasas de resistencia a tetraciclina (38,2% por CLSI y 50,9% por EUCAST) y ciprofloxacino (49,1% CLSI y 54% EUCAST), y bajos porcentajes de resistencia a espectinomicina (0%), cefixima (0,5% CLSI pero 5,9% EUCAST) y ceftriaxona (1,5% CLSI y 2,4% EUCAST). La resistencia a azitromicina fue del 6% (tanto CLSI como EUCAST). El análisis molecular reveló la presencia de 86 secuenciotipos (ST) distintos, destacando el ST2992 (7,4%), ST3378 (6,9%), ST2400 (4,6%) y ST13288 (6,9%) el cual presentaba asociación con resistencia a cefixima (P=,031). Los genogrupos (G) mayoritarios fueron el G1407 (13,1%), G2992 (10,3%), G2400 (6,3%) y G387 (3,4%); G1407 y G2400 mostraron asociación con resistencia a ciprofloxacino (P<,03).
ConclusiónSe ha detectado una baja resistencia a ceftriaxona, una preocupante resistencia a azitromicina y una gran variedad de secuenciotipos circulantes, algunos de los cuales presentan correlación con determinados perfiles de resistencia.
Infections caused by Neisseria gonorrhoeae (NG) represent a serious public health problem1. In Europe, the data reported by the European Centre for Disease Prevention and Control (ECDC) point to a marked and progressively increasing incidence2. Prompt diagnosis and appropriate treatment combined with preventive and educational measures are fundamental factors in the control of gonococcal infection (GI). In 2012, the main scientific societies started recommending empirical dual therapy of gonorrhoea by combining a third-generation cephalosporin, usually ceftriaxone, with a second-generation macrolide, azithromycin. The appearance in 2018 of the first strain resistant to this combination, along with the change in patterns of susceptibility to antimicrobials, led to the need to reassess the treatment regimen3–5.
Molecular epidemiology is an essential tool for understanding population dynamics6, as it enables relationships to be established between local antibiotic susceptibility patterns and circulating strains7, which makes it very useful in identifying strains with high resistance and their possible spread among the population8. The constant increase in GI cases worldwide, the lack of effective vaccines and the emergence of strains resistant to a wide variety of potential therapeutic options (the World Health Organisation [WHO] recently classified it as a superbug9) underline the need to implement surveillance and infection control programmes.
The main aim of the GONOvig project was to analyse the antibiotic susceptibility of NG strains isolated in genital samples from patients in the Valencian Community, as well as to carry out molecular typing of the isolates in order to detect epidemiological clusters and their possible association with the resistance profiles detected.
MethodsNG isolatesWe conducted a prospective, multicentre study, in which we analysed 227 NG strains obtained from urethral or endocervical swabs taken from patients treated in thirteen representative hospitals in the Valencian Community from January 2013 to December 2018. In order to have a certain geographical representation, a minimum of five isolates were required for each participating centre, corresponding to a single strain per patient and episode. Samples considered to be from sites of colonisation (pharyngeal or anal exudates) were not included in the study, due to the lower prevalence of NG in these biological specimens. Each participating centre identified and carried out the antibiotic susceptibility testing of the isolates using methods available in their laboratory. These centres then sent the original swab and primary culture, refrigerated at 2−8°C, to Hospital Universitario de La Ribera (HULR), no later than 24−48h after identifying the isolate, along with demographic data on the patient and the antibiotic susceptibility test carried out at source. The HULR, a reference centre, re-spread the isolates on Chocolate agar and Thayer-Martin selective medium (bioMerieux, Marcy-l'Étoile, France), and the isolates were re-identified based on the morphology of the colonies, a positive result in the tests for oxidase and catalase, and a high degree of reliability (>95%) in the biochemical gallery using the Vitek-2 system10 (bioMerieux, Marcy-l'Étoile, France).
Antibiotic susceptibility testsThe isolates collected during the GONOvig project were analysed following the international guidelines of the Clinical and Laboratory Standards Institute (CLSI)11 and European Committee on Antimicrobial Susceptibility Testing (EUCAST)12. ETEST strips (bioMerieux, Marcy-l'Étoile, France) were used and the Minimum Inhibitory Concentration (MIC) was determined in GC agar, supplemented with 1% IsoVitaleX (BD, New Jersey, United States) and incubated for 20−24h in the oven at 37°C with 5% CO2, against the following antibiotics: penicillin, cefixime, ceftriaxone, azithromycin, ciprofloxacin, spectinomycin and tetracycline. As quality control, the strain NG ATCC 49226 was used.
To minimise possible discrepancies in the reading of the inhibition ellipses, the MIC value was determined by two specialists. The interpretation was carried out comparatively according to CLSI and EUCAST criteria. Additionally, the MIC capable of inhibiting 50% (MIC50) and 90% (MIC90) of the isolates was calculated. We also studied β-Lactamase (penicillinase-producing Neisseria gonorrhoeae [PPNG]) production using the cefinase test (BD, New Jersey, USA).
Molecular typingMolecular characterisation was centralised at the Valencian Community's Fundación para el Fomento de la Investigación Sanitaria y Biomédica (FISABIO) [Foundation for the Promotion of Health and Biomedical Research]. The NG multi-antigen sequence typing (NG-MAST) technique was used, based on the sequencing of internal fragments of the hypervariable porB and tbpB genes, which encode the porin and transferrin of the outer membrane of NG. The QIAamp® DNA Mini Kit system (Qiagen, Hilden, Germany) was used. Allele sequences were amplified according to the protocol already described13, using a Mastercycler® (Eppendorf, Hamburg, Germany) thermal cycler, Master Mix reagents (Biotools, Madrid, Spain) and specific primers (Metabion, Munich, Germany). The allele sequences obtained were entered into the database (www.ng-mast.net) and the resulting sequence types (ST) were recorded. The ST obtained were then grouped into larger associations, called genogroups (G), based on allele similarities14. To achieve this, the sequences were aligned and associations were established using the MEGA program (https://www.megasoftware.net).
Statistical analysis of the dataQuantitative variables are presented as mean and standard deviation. The Anderson-Darling test was used to verify the normality of the distribution of the quantitative variables, P<.05 being considered as statistically significant, with a confidence level of 95%. To establish relationships between qualitative variables, such as antibiotic susceptibility and ST/genogroups, contingency tables were created and Fisher's exact test applied. The statistical calculations were performed with the SPSS Statistics 23 program (IBM, Armonk, United States).
ResultsFor the strains, 100% were re-identified as NG at the reference centre. Of the 227 isolates analysed, 70% came from hospitals in the city of Valencia, 15% from Alicante and 15% from Castellón. By gender, 216 of the isolates were obtained in males (94%) and 11 in females (6%). The mean age of the population studied was 30.7 (±9.4) years, with a range from 17 to 79.
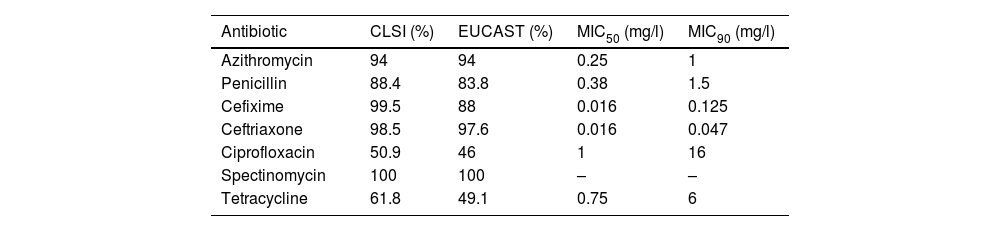
Table 1 shows the main results obtained after the antibiotic susceptibility studies. Of the patients with isolates, 6% had MIC>1mg/l to azithromycin and were classified as non-wild-type phenotypes carrying mechanisms of resistance15. The proportion of strains categorised as susceptible to penicillin was 4.1% (both in CLSI and EUCAST), while 84.3% (CLSI) and 79.7% (EUCAST) were of intermediate susceptibility, and 11.6% (CLSI) and 16.2% (EUCAST) were resistant. Of the gonococci, 5.6% showed plasmid resistance to penicillin (PPNG). The proportion of strains resistant to third-generation cephalosporins was 0.5% (CLSI) and 5.9% (EUCAST) for cefixime, and 1.5% (CLSI) and 2.4% (EUCAST) for ceftriaxone. Approximately 50% of the isolates were resistant to ciprofloxacin (CLSI and EUCAST), and 38.2% (CLSI) and 50.9% (EUCAST) were resistant to tetracycline. All the strains analysed were susceptible to spectinomycin.
Percentages of susceptibility to various antibiotics in Neisseria gonorrhoeae isolates: comparison according to CLSI and EUCAST criteria.
| Antibiotic | CLSI (%) | EUCAST (%) | MIC50 (mg/l) | MIC90 (mg/l) |
|---|---|---|---|---|
| Azithromycin | 94 | 94 | 0.25 | 1 |
| Penicillin | 88.4 | 83.8 | 0.38 | 1.5 |
| Cefixime | 99.5 | 88 | 0.016 | 0.125 |
| Ceftriaxone | 98.5 | 97.6 | 0.016 | 0.047 |
| Ciprofloxacin | 50.9 | 46 | 1 | 16 |
| Spectinomycin | 100 | 100 | – | – |
| Tetracycline | 61.8 | 49.1 | 0.75 | 6 |
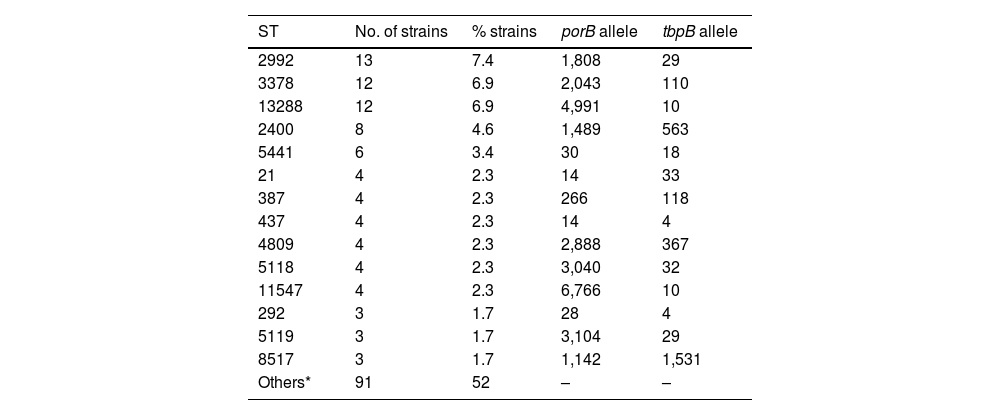
For technical and budgetary reasons, molecular characterisation was carried out on 175 strains (77% of the total). These typing studies revealed the existence of 86 different ST, made up of 73 different porB alleles and 36 different tbpB alleles. Nine isolates could not be ST-classified because they corresponded to new sequences or allele combinations not included in the NG-MAST database. The most prevalent ST were ST2992 (n=13), ST3378 (n=12), ST13288 (n=12), ST2400 (n=8) and ST5441 (n=6). The ST detected in ≥3 isolates and their alleles are listed in Table 2.
Main sequence types detected and constituent alleles (porB and tbpB).
| ST | No. of strains | % strains | porB allele | tbpB allele |
|---|---|---|---|---|
| 2992 | 13 | 7.4 | 1,808 | 29 |
| 3378 | 12 | 6.9 | 2,043 | 110 |
| 13288 | 12 | 6.9 | 4,991 | 10 |
| 2400 | 8 | 4.6 | 1,489 | 563 |
| 5441 | 6 | 3.4 | 30 | 18 |
| 21 | 4 | 2.3 | 14 | 33 |
| 387 | 4 | 2.3 | 266 | 118 |
| 437 | 4 | 2.3 | 14 | 4 |
| 4809 | 4 | 2.3 | 2,888 | 367 |
| 5118 | 4 | 2.3 | 3,040 | 32 |
| 11547 | 4 | 2.3 | 6,766 | 10 |
| 292 | 3 | 1.7 | 28 | 4 |
| 5119 | 3 | 1.7 | 3,104 | 29 |
| 8517 | 3 | 1.7 | 1,142 | 1,531 |
| Others* | 91 | 52 | – | – |
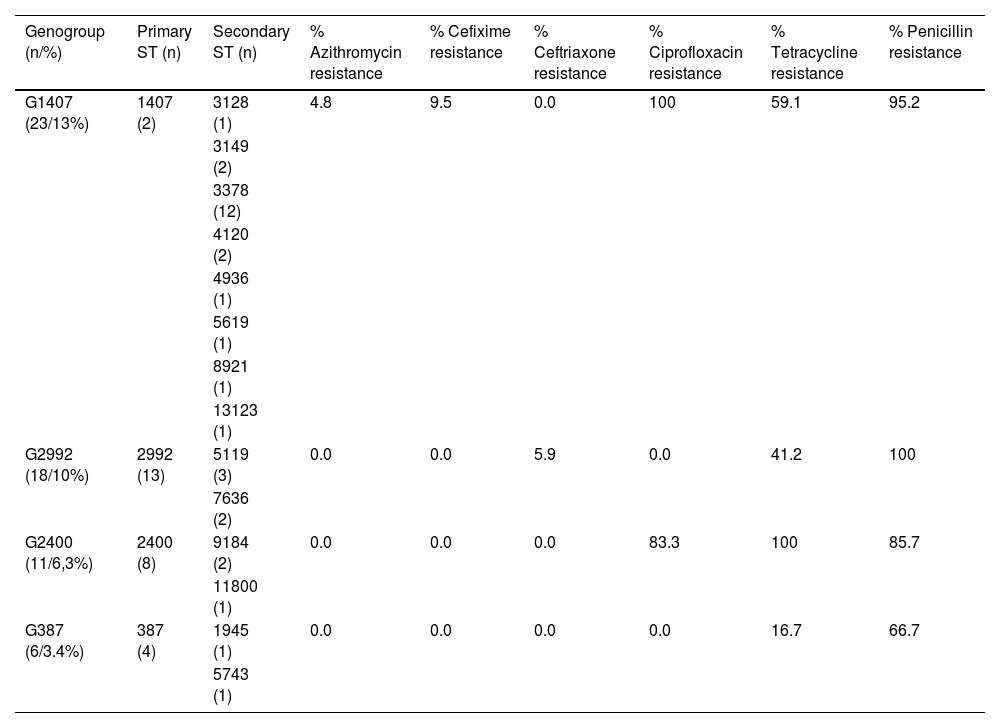
Table 3 shows the four most identified genogroups, along with their ST (primary and secondary) and percentages of resistance associated with antibiotics (according to EUCAST criteria); G1407 (13.1%), followed by G2992 (10.3%), G2400 (6.3%) and G387 (3.4%). The 23 strains included in the main genogroup, G1407, showed resistance to ciprofloxacin (P>.02), this genogroup being made up mainly of 13 isolates of ST3378 and only two with ST1407. Two isolates (ST1407 and ST3378) were resistant to cefixime (MIC 0.19mg/l in both cases) and a third isolate (ST3378) had an MIC>1mg/l against azithromycin. One strain belonging to G2992 (ST2992) was resistant to ceftriaxone (MIC 0.75mg/l). G2400, primarily comprising strains with ST2400, was associated with resistance to ciprofloxacin (P=.017) and tetracycline (P=.048).
The most prevalent genogroups and percentages of associated antibiotic resistance.
| Genogroup (n/%) | Primary ST (n) | Secondary ST (n) | % Azithromycin resistance | % Cefixime resistance | % Ceftriaxone resistance | % Ciprofloxacin resistance | % Tetracycline resistance | % Penicillin resistance |
|---|---|---|---|---|---|---|---|---|
| G1407 (23/13%) | 1407 (2) | 3128 (1) | 4.8 | 9.5 | 0.0 | 100 | 59.1 | 95.2 |
| 3149 (2) | ||||||||
| 3378 (12) | ||||||||
| 4120 (2) | ||||||||
| 4936 (1) | ||||||||
| 5619 (1) | ||||||||
| 8921 (1) | ||||||||
| 13123 (1) | ||||||||
| G2992 (18/10%) | 2992 (13) | 5119 (3) | 0.0 | 0.0 | 5.9 | 0.0 | 41.2 | 100 |
| 7636 (2) | ||||||||
| G2400 (11/6,3%) | 2400 (8) | 9184 (2) | 0.0 | 0.0 | 0.0 | 83.3 | 100 | 85.7 |
| 11800 (1) | ||||||||
| G387 (6/3.4%) | 387 (4) | 1945 (1) | 0.0 | 0.0 | 0.0 | 0.0 | 16.7 | 66.7 |
| 5743 (1) |
It is estimated that more than 376 million new sexually transmitted infections (STI) occur each year, representing a serious public health problem worldwide. GI is the second most prevalent STI of bacterial aetiology, causing a total of 86 million new infections in 2016 alone16, a significantly higher figure than the 78 million reported by the WHO in 2012. In addition to the increase in its incidence, NG has shown an exceptional capacity to develop mechanisms enabling it to evade the action of many of the antimicrobial agents used for its eradication. The appearance of strains resistant to azithromycin15,17 and the changes in the resistance profile of NG have led to modifications to the drug treatment regimen. The current recommendations of the American Centers for Disease Control and Prevention (CDC)3, British Association for Sexual Health and HIV (BSHH)5 and European4 guidelines propose doubling the dose of ceftriaxone and, in the case of the CDC and BSHH, abandoning the use of azithromycin, establishing ceftriaxone monotherapy as the most recommended empirical therapy.
The 2017 report of the European Gonococcal Antimicrobial Surveillance Programme (Euro-GASP) highlights Spain as one of the countries with the highest rates of resistance to oral cefixime (5.5%) in the period 2009–201718, a finding consistent with our analysis (5.9%). With regards to Spain, in 2011 Pladevall analysed strains isolated in Barcelona that showed resistance rates of 10% to cefixime and 3% to ceftriaxone19. A subsequent study of the same group with a greater number of strains determined a resistance of 8.3% to cefixime and 2.8% to ceftriaxone20. Cobo et al. reported resistance rates of 6.1% to cefixime and 4.6% to ceftriaxone after analysing samples collected in southern Spain21, which decreased to 3.7% (cefixime) and 2.9% (ceftriaxone) in subsequent studies22. The study carried out in Valencia between 2013 and 2019 by Castaño et al. detected 3% of isolates resistant to cefixime and 0.4% to ceftriaxone23.
Despite the slight geographical and temporal discrepancies, these data confirm NG resistance close to 5% for cefixime and 2% for ceftriaxone according to EUCAST criteria. However, when applying CLSI criteria, these rates are lower, with a particularly evident discrepancy for cefixime (0.5% vs 5.9%), as a result of the different breakpoints established by the two institutions. This can cause problems when classifying the isolates, making it desirable to standardise the cut-off values based on clinical, microbiological and pharmacokinetic criteria.
In the case of azithromycin, an antimicrobial considered the cornerstone in the treatment of several STI, both societies establish an epidemiological cut-off value (ECV) of 1mg/l, which is the MIC that differentiates wild-type strains11,12. Modern studies suggest that NG has acquired chromosomal mutations in 23S rRNA in recent years15 which, in conjunction with other resistance mechanisms, enable it to evade the action of macrolides. In Europe, the ECDC24 reported 7.6% resistance to azithromycin in 2018, representing a significant increase compared to the 3.7% in 2017. Resistance to azithromycin detected in other national studies in Spain22,23 has been 9% and 9.7%.
The molecular characterisation of the isolates revealed that most (59.3%) of the 86 ST identified were present in a single strain. The main ST detected in our study have also been identified at a European level14,25, with the exception of ST13288, in which three strains with resistance to cefixime were included, alerting us to the possible appearance of a clone, only reported in Italy26, with the potential ability to evade the empirical therapy of choice in GI.
According to the scientific literature, the main genogroup in Spain and Europe is G1407, closely linked to therapeutic failure in France27, Spain28 and the United Kingdom29, as well as other countries. In our analysis, its high prevalence was mainly defined by the 12 strains belonging to ST3378, one of which was resistant to cefixime and another to azithromycin, as well as a statistically significant association with ciprofloxacin-resistant isolates (P=.02).
Chisholm et al. performed the molecular typing of 1066 NG strains obtained in 21 European countries participating in the European Gonococcal Antimicrobial Surveillance Programme (Euro-GASP) from 2009 to 201014 concluding that, here in Spain, the most prevalent was G1407, followed by G2992, in line with the results obtained in our study. The ECDC also reported that G1407 was the most common genogroup in Europe in 2009 (23.3%), with its incidence decreasing in 2013 (14.8%)26, followed by genogroups G2992 and G2400, which showed association with strains resistant to ciprofloxacin (P=.017) and tetracycline (P=.048). The increase in G2400 is a finding also verified by Pladevall, along with its facility for developing antimicrobial resistance mechanisms20. Lastly, in our analysis, G387 had rates similar to those reported in the above European studies14,26.
Establishing links between ST and/or genogroups and specific resistance phenotypes requires molecular typing analyses alongside antibiotic susceptibility testing of the isolates. This highlights the utility of the NG-MAST molecular technique as a complementary method in understanding antimicrobial resistance. Conventional culture continues to be essential to determine the in vitro antibiotic susceptibility profile as soon as possible and establish the most appropriate treatment in each case.
One limitation of our study is the fact that the strains analysed belonged only to patients in the Valencian Community. Surveillance, both epidemiological and molecular, is required in areas and countries around us in order to compare results. Another limitation was, despite their lower prevalence, not having included isolates from pharyngeal and anal samples; these have been reported to potentially have different resistance profiles, as the colonising strains in these anatomical areas are subject to less antibiotic pressure and a greater acquisition of resistance genes from other saprophytic species of NG30. Moreover, we did not collect information on the sexual orientation of the patients included in the GONOvig project, and that could be of interest when establishing clinical-epidemiological associations.
In conclusion, the molecular data we obtained show that the circulating NG population in the Valencian Community has similarities with strains isolated in other geographical areas, with ST2992 and ST2400 found to have a particularly high prevalence. Our research shows that there is a correlation between certain ST and/or genogroups and antibiotic resistance, especially to ciprofloxacin and tetracycline. Last of all, resistance to third-generation cephalosporins remained low, particularly for ceftriaxone (2.4%) compared to cefixime (5.9%), while strains with high MIC of azithromycin have increased in recent years (6%).
FundingThe GONOvig project received two research support grants from the Hospital Universitario de La Ribera (HULR). These grants were used to purchase the material and means necessary to carry out the experimental phase of the study.
Conflicts of interestThe authors declare that they have no conflicts of interest.
We would like to thank the Fundación para el Fomento de la Investigación Sanitaria y Biomédica (FISABIO) [Foundation for the Promotion of Health and Biomedical Research], as well as the technical and medical staff who collaborated in the GONOvig Project.