To describe the bacterial profile of the supragingival biofilm of children with temporary dentition (CTD) and early mixed dentition (CEMD), with the next-generation sequencing (HOMINGS) technique.
MethodA comparative descriptive study was carried out with 30 systemically healthy children aged between 5 and 7 years old from public schools in Cartagena-Colombia. All participants were caries-free applying the criteria of the International Caries Detection and Assessment System (ICDAS II) and had no caries experience according to the Decayed, Missing and Filled Teeth (DMFT) index. Supragingival biofilm samples were collected. Bacterial DNA was extracted and used for analysis using HOMINGS (Human Oral Microbe Identification using Next-Generation Sequencing) based on the sequencing of the V3–V4 region of the 16S rRNA gene using the Illumina MiSeq platform (V3–V4 primers).
ResultsA total of 360 species-specific and 65 genus-specific probes were identified. The bacterial genus most predominant in CTD were Streptococcus, Actinomyces, Veillonella and Fusobacterium (29.2% of all bacterial DNA present), while in CEMD the most predominant were Streptococcus, Leptotrichia, TM7 and Porphyromonas (24.5% of all bacterial DNA present). The bacterial species with the highest relative abundance in the oral biofilm microbiome from CTD were Streptococcus sanguinis, Rothia aeria, Gemella haemolysans, while in CEMD they were S. sanguinis, Leptotrichia spp. HOT-417 and Leptotrichia spp. HOT-498. The Shannon diversity index was 2.77 (SD=0.26) for CTD and 3.01 (SD=0.39) for CEMD (p=0.06).
ConclusionsThe analysis of the bacterial profile of the supragingival dental biofilm in children with DMFT, by means of HOMINGS showed low microbiological diversity both in presence and in relative abundance in terms of genus as well as bacterial species.
Describir el perfil bacteriano del biofilm supragingival de niños con dentición temporal (NDT) y dentición mixta temprana (NDMT), con la técnica de secuenciación de próxima generación HOMINGS.
MétodoSe realizó un estudio descriptivo comparativo con 30 niños de 5 a 7 años de edad sistémicamente sanos de escuelas públicas de Cartagena (Colombia). Todos los participantes estaban libres de caries, según los criterios del Sistema Internacional de Detección y Evaluación de Caries (ICDAS II) y sin experiencia de caries según el índice de dientes cariados, perdidos y obturados (DCPO). Se recolectaron muestras de biofilm supragingival. Se extrajo el ADN bacteriano y se usó para su análisis mediante HOMINGS (identificación de microorganismos orales humanos utilizando secuenciación de próxima generación) basado en la secuenciación de la región V3-V4 del gen 16S rRNA con la plataforma Illumina MiSeq.
ResultadosSe identificaron 360 especies específicas y 65 géneros específicos de las sondas: Streptococcus, Actinomyces, Veillonella y Fusobacterium (29,2% del total de ADN bacteriano presente), mientras que en el grupo de dentición mixta temprana se encontraban Streptococcus, Leptotrichia, TM7 y Porphyromonas (24,5% del ADN bacteriano presente). Las especies bacterianas con mayor abundancia relativa en el microbioma oral de biofilm de NDT fueron Streptococcus sanguinis, Rothia aeria, Gemella haemolysans, mientras que en NDMT fueron S. sanguinis, Leptotrichia sp. HOT-417, Leptotrichia sp. HOT-498. El índice de diversidad de Shannon fue 2,77 (DE=0,26) para NDT y 3,01 (DE=0,39) para NDMT (p=0,06).
ConclusionesEl análisis del perfil bacteriano del biofilm dental supragingival en niños con NDMT mediante HOMINGS mostró baja diversidad microbiológica tanto en presencia como en abundancia relativa a nivel de género y de especies bacterianas.
The human microbiome plays an important role in health and well-being. In recent years, a great deal of work has been done to characterise the different microbial communities that colonise the human body, and of all the different sites, the mouth houses one of the most diverse communities.1,2
With the advent of molecular techniques for the identification of oral bacteria based on DNA, it has been shown that the oral microbiota is much more complex and diverse than originally thought.3 The mouth is colonised by over 700 species of microorganisms, 35% of which are still unculturable. There may be a difference between this microbial community in childhood and adulthood.4
Biofilms are heterogeneous accumulations of a microbial community, surrounded by an intercellular matrix of polymers. These microorganisms can adhere to or deposit themselves on many sites, including the walls of the teeth, the oral mucosa and the saliva.5 Some examples of tooth colonisers which contribute to the formation of the oral biofilm are Streptococcus sanguis, S. mitis and S. oralis, Actinomyces naeslundii, S. mutans, S. salivarius, S. gordonii, S. parasanguis, Neisseria spp., Prevotella loescheii, P. intermedia, Capnocytophaga spp., Fusobacterium nucleatum and Porphyromonas gingivalis.6,7
The recently developed Human Oral Microbe Identification using Next-Generation Sequencing (HOMINGS) technique enables the simultaneous species identification of nearly 600 oral bacterial taxa. Advantages of the technique include the fact that it is computationally efficient, rapid and reproducible, and it can identify the species of most of the oral microbiome.8 HOMINGS has been used in several recent research studies. In 2017, Richards et al. investigated the microbiome of dental plaque in children with different states of caries9 and in 2016, Belstrom et al. studied the temporal stability of salivary microbiomes.10
The purpose of this study was to identify the bacterial profile of the dental biofilm in children with deciduous and early mixed dentition using the HOMINGS technique, as this technology would provide more complete information about the supragingival microbiome.
MethodsDescriptive cross-sectional study model in which the bacterial profile of the oral biofilm was described in children with deciduous dentition and early mixed dentition. The study population was children from five to seven years of age, from three primary schools in the city of Cartagena de Indias (Colombia). The sample size was determined using statistical software Epidat v. 4.0. For this, we used an expected caries prevalence in the Caribbean region of 90%, as reported in ENSAB IV (IV Estudio nacional de salud bucal [4th Colombian Study of Oral Health]),11 with a confidence interval of 95% and error of 5%. The sample was made up of 30 children.
We excluded children with systemic, haematological or autoimmune disease, children on pharmacological treatment in the three months prior to the start of the study, children with congenital malformations involving the oral cavity, children whose last visit to the dentist was within the previous six months who received dental prophylaxis, sealants and/or topical applications of fluoride gel or varnish during that time, as these procedures can change the microenvironment of the mouth.
This project was developed as part of the project “Characterisation of the oral microbiome in saliva and bacterial biofilm of children with caries. Prospective case–control study”, code N°. 141965741160, which was funded by Colciencias (contract 673-2014) and carried out by Corporación Universitaria Rafael Núñez. The activities described here, the study sample and the funding are completely derived from the aforementioned project.
Techniques and procedures for data collectionSamples of supragingival biofilm were collected from healthy children from the first permanent molar in children with early mixed dentition and from the second deciduous molar in children with deciduous dentition. In cases where the tooth did not have supragingival biofilm, plaque was collected from nearby surfaces. The supragingival biofilm was collected with an applicator and placed in 1.5-ml tubes with 150μl of TE buffer and stored at −20°C until further analysis.
Extraction of microbial DNAFor the extraction of bacterial DNA, the samples (biofilm) were suspended in 150μl of TE buffer (10mM Tris–HCl, 1M EDTA, pH 8.0) and 1μl of Ready-Lyse Lysozyme (Epicentre Cat. No. R1802M, USA), and incubated at 37°C overnight. Next, 150μl of 2× T and C lysis solution and 1μl of proteinase K were added and it was incubated for 30min at 65°C with continuous shaking.
The samples were placed on ice for 3–5min and then 175μl of MPC Protein Precipitation Reagent were added to 300μl of the lysed samples; they were shaken vigorously for 10s and then centrifuged for 10min at 4°C at 10,000×g. The supernatant was transferred to 1.5-ml sterile tubes, 500μl of isopropanol were added and the tubes were mixed 30–40 times. They were incubated for 10min on ice, centrifuged at 10,000×g at 4°C and then the isopropanol discarded, avoiding any loss of precipitate. The precipitate was rinsed with 500μl of 75% ethanol, mixed, briefly centrifuged and the ethanol discarded; this step was then repeated. The DNA was resuspended with 25μl of TE buffer. Each sample was analysed for DNA concentration and quality using a Multiskan Go® spectrophotometer (ThermoFisher Scientific) at low optical densities of 260/280. The samples were stored at −20°C until further analysis.
Bacteria identificationThe microbiological profile obtained from the purified DNA of the biofilm samples was made using the HOMINGS technology (http://homings.forsyth.org). HOMINGS is a molecular technique based on next-generation sequencing using the MiSeq System platform (Ilumina, Inc., San Diego, CA, USA), which sequences the region 16S rRNA V3–V4 using described primers.12 This technique was developed at the Forsyth Institute (Boston, MA, USA) and, in conjunction with a customised BLAST programme, ProbeSeq programme for HOMINGS (Forsyth Institute, Cambridge, MA, USA), is able to simultaneously identify around 600 oral taxa.12 The ProbeSeq database contains sequences of probes (17–40 bases) collected in the HOMD (Human Oral Microbiome Database) database; in particular, 598 species-specific probes and 94 genus-specific probes (probes targeting two or more closely related taxa within the same genus).
Statistical analysisThe sociodemographic data such as gender, schooling and presence and relative abundance of the organism are shown in frequency tables. The age variable is expressed as mean and standard deviation (SD). The comparison between demographic variables and type of dentition was analysed using Pearson's chi-square test. Shannon's α microbiological diversity index was estimated from the number of readings obtained per sample. Student's t test was used to compare the Shannon Index between the study groups. A p-value <0.05 was considered significant for these analyses.
The relative abundance and presence of bacterial species and genera did not show normal distribution according to the Shapiro–Wilk test of normality. The comparison of these variables between the groups according to the type of dentition was carried out using the Mann–Whitney U test with Benjamini–Hochberg correction for multiple comparisons. This was used to control the discovery of false positives of 10% (66). For these analyses, an adjusted p-value <0.001 was considered statistically significant. Statistical analyses were performed with the statistical programmes SPSS v20 (IBM, New York, USA) and MeV 4.8.1 MultiExperiment Viewer; www.tm4.org.13,14
Ethical considerationsBefore initiating any procedure, the parents or legal guardians approved the participation of the minor through an informed consent form. The child was asked about their willingness to take part in the study using an assent form. According to resolution 008430 of 1993, this study represents a greater-than-minimal risk for the participants: the possible risks were clearly explained in the informed consent form.
ResultsThirty children took part in the study: 15 with deciduous dentition and 15 with early mixed dentition. The mean age was 6.23±0.8 years, 46.7% (n=14) were male and 53.3% (n=16) were female. They were enrolled in three primary schools in the ‘Localidad 1 Histórica y del Caribe Norte’ district in the city of Cartagena (Bolívar, Colombia).
The HOMINGS analysis enabled identification of the bacterial profile in samples of oral biofilm from supragingival surfaces, and revealed the bacterial diversity of some of the bacterial species identified and their contribution (relative proportion or abundance) to the microbiome of each individual. In total, 65 genus-specific probes were amplified in the study samples; these collected information from 44 bacterial genera present in the oral biofilm analysed. Of these, the genera Streptococcus, Actinomyces, Veillonella, TM7, Porphyromonas, Fusobacterium, Neisseria, Rothia, Selenomonas and Centipeda, Leptotrichia, Gemella, Actinomyces and Corynebacterium were present in 100% of the samples. The relative abundance (relative proportion) of each of the genera denotes their contribution to the bacterial profile of the oral biofilm. Streptococcus probe 4 (20.67%±12.1), Streptococcus probe 1 (2.68%±2.67), Actinomyces probe 3 (1.07%±1.3), Leptotrichia probe 3 (0.96%±1.7), Veillonella probe 2 (0.91%±1.4), TM7 (0.90%±2.0) and Porphyromonas probe 2 (0.82%±1.3) were the six most abundant bacterial genera in the oral biofilm, representing 28.02% of the biofilm.
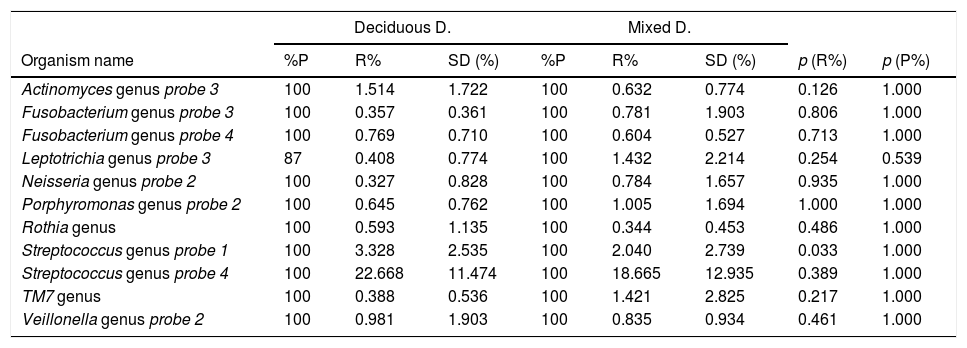
Bacterial profile of oral biofilm according to the type of dentitionThe bacterial profile of the oral biofilm varied between the two types of dentition, both in terms of presence of and relative abundance in the genus, and in the bacterial species. In the bacterial genera, it was found that the bacteria with the highest relative abundance in the biofilm samples from children with deciduous dentition were Streptococcus probe 4, Streptococcus probe 1, Actinomyces genus probe 3, Veillonella probe 2, Fusobacterium probe 4 and Porphyromonas genus probe 2. In contrast, in the children with mixed dentition we found Streptococcus probe 4, Streptococcus probe 1, Leptotrichia probe 3, TM7 genus, Porphyromonas genus probe 2, Veillonella genus probe 2 and Neisseria genus probe 2 (Table 1).
Bacteria genera according to type of dentition.
| Deciduous D. | Mixed D. | |||||||
|---|---|---|---|---|---|---|---|---|
| Organism name | %P | R% | SD (%) | %P | R% | SD (%) | p (R%) | p (P%) |
| Actinomyces genus probe 3 | 100 | 1.514 | 1.722 | 100 | 0.632 | 0.774 | 0.126 | 1.000 |
| Fusobacterium genus probe 3 | 100 | 0.357 | 0.361 | 100 | 0.781 | 1.903 | 0.806 | 1.000 |
| Fusobacterium genus probe 4 | 100 | 0.769 | 0.710 | 100 | 0.604 | 0.527 | 0.713 | 1.000 |
| Leptotrichia genus probe 3 | 87 | 0.408 | 0.774 | 100 | 1.432 | 2.214 | 0.254 | 0.539 |
| Neisseria genus probe 2 | 100 | 0.327 | 0.828 | 100 | 0.784 | 1.657 | 0.935 | 1.000 |
| Porphyromonas genus probe 2 | 100 | 0.645 | 0.762 | 100 | 1.005 | 1.694 | 1.000 | 1.000 |
| Rothia genus | 100 | 0.593 | 1.135 | 100 | 0.344 | 0.453 | 0.486 | 1.000 |
| Streptococcus genus probe 1 | 100 | 3.328 | 2.535 | 100 | 2.040 | 2.739 | 0.033 | 1.000 |
| Streptococcus genus probe 4 | 100 | 22.668 | 11.474 | 100 | 18.665 | 12.935 | 0.389 | 1.000 |
| TM7 genus | 100 | 0.388 | 0.536 | 100 | 1.421 | 2.825 | 0.217 | 1.000 |
| Veillonella genus probe 2 | 100 | 0.981 | 1.903 | 100 | 0.835 | 0.934 | 0.461 | 1.000 |
p<0.001. Mann–Whitney U test with Benjamini–Hochberg correction (false discovery rate: 10%) of the multiple comparisons. Bacteria genera are shown with an average relative proportion ≥0.3% in the microbiome of the individuals.
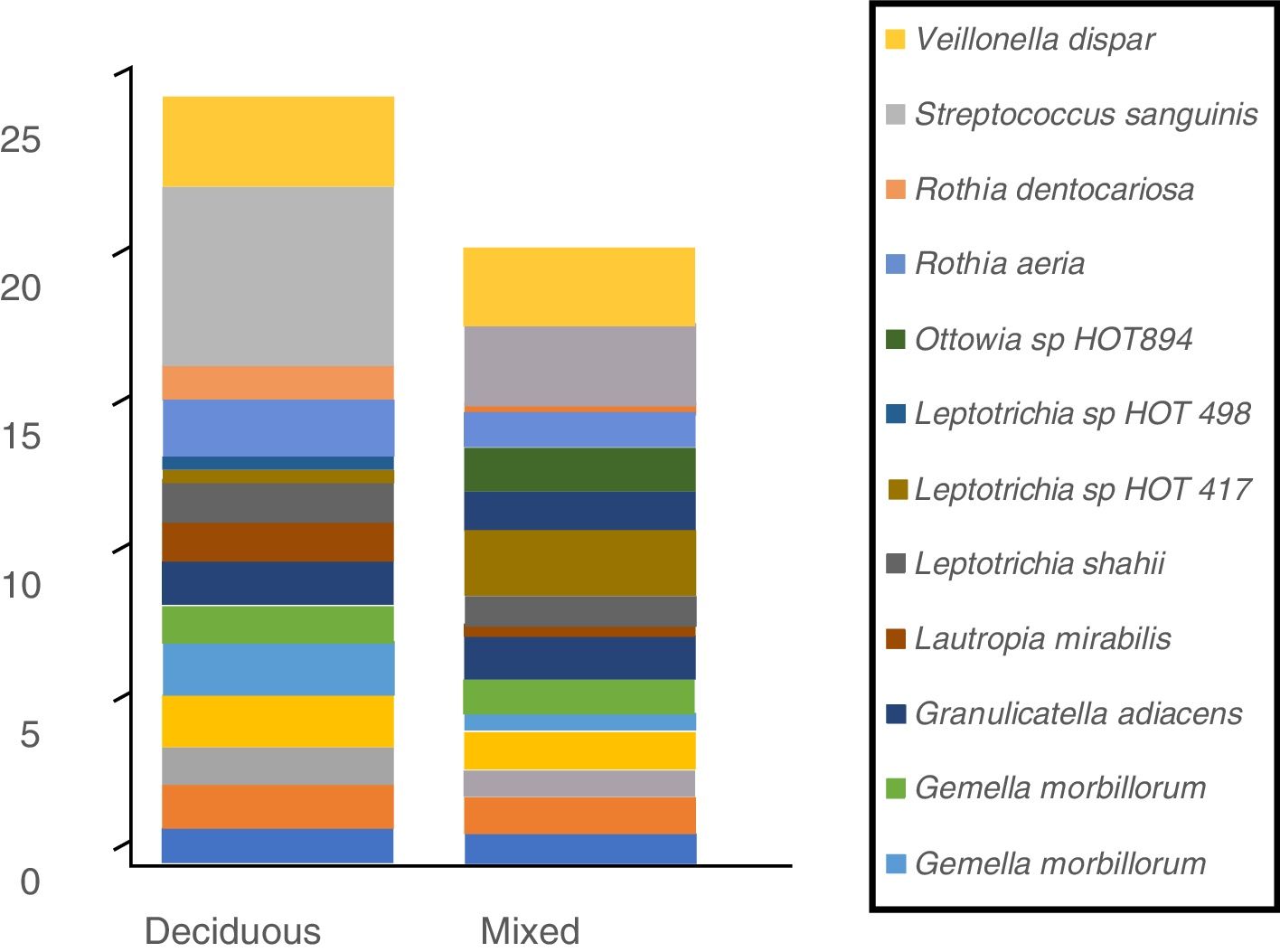
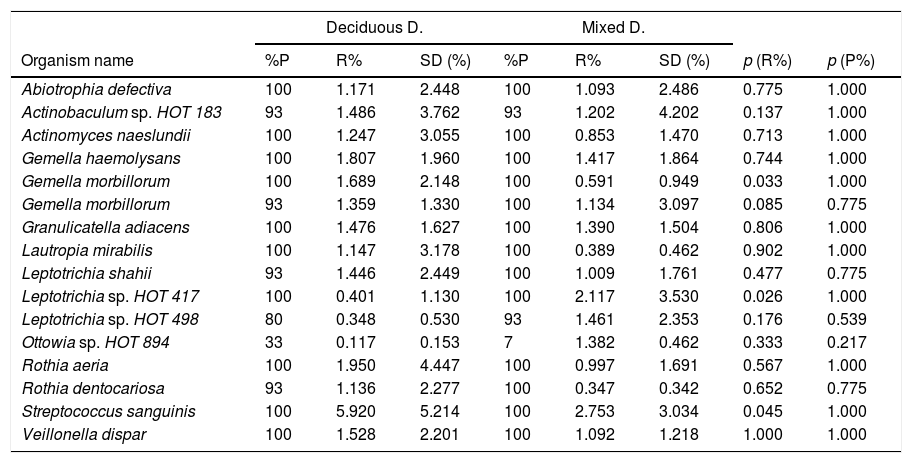
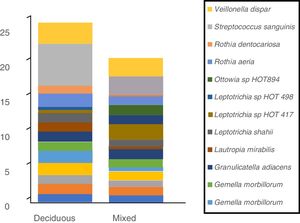
In terms of the bacterial species, the six species with the highest relative abundance in the biofilm samples from children with deciduous dentition were: Streptococcus sanguinis, Rothia aeria, Gemella haemolysans, Gemella morbillorum, Veillonella dispar and Actinobaculum sp. HOT 183. While in children with early mixed dentition the most prevalent bacterial species were: Streptococcus sanguinis, Leptotrichia sp. HOT 417, Leptotrichia sp. HOT 498, Gemella haemolysans, Granulicatella adiacens and Ottowia sp. HOT 894 (Table 2). Fig. 1 shows the bacterial species with the highest relative abundance.
Microorganism species according to dentition.
| Deciduous D. | Mixed D. | |||||||
|---|---|---|---|---|---|---|---|---|
| Organism name | %P | R% | SD (%) | %P | R% | SD (%) | p (R%) | p (P%) |
| Abiotrophia defectiva | 100 | 1.171 | 2.448 | 100 | 1.093 | 2.486 | 0.775 | 1.000 |
| Actinobaculum sp. HOT 183 | 93 | 1.486 | 3.762 | 93 | 1.202 | 4.202 | 0.137 | 1.000 |
| Actinomyces naeslundii | 100 | 1.247 | 3.055 | 100 | 0.853 | 1.470 | 0.713 | 1.000 |
| Gemella haemolysans | 100 | 1.807 | 1.960 | 100 | 1.417 | 1.864 | 0.744 | 1.000 |
| Gemella morbillorum | 100 | 1.689 | 2.148 | 100 | 0.591 | 0.949 | 0.033 | 1.000 |
| Gemella morbillorum | 93 | 1.359 | 1.330 | 100 | 1.134 | 3.097 | 0.085 | 0.775 |
| Granulicatella adiacens | 100 | 1.476 | 1.627 | 100 | 1.390 | 1.504 | 0.806 | 1.000 |
| Lautropia mirabilis | 100 | 1.147 | 3.178 | 100 | 0.389 | 0.462 | 0.902 | 1.000 |
| Leptotrichia shahii | 93 | 1.446 | 2.449 | 100 | 1.009 | 1.761 | 0.477 | 0.775 |
| Leptotrichia sp. HOT 417 | 100 | 0.401 | 1.130 | 100 | 2.117 | 3.530 | 0.026 | 1.000 |
| Leptotrichia sp. HOT 498 | 80 | 0.348 | 0.530 | 93 | 1.461 | 2.353 | 0.176 | 0.539 |
| Ottowia sp. HOT 894 | 33 | 0.117 | 0.153 | 7 | 1.382 | 0.462 | 0.333 | 0.217 |
| Rothia aeria | 100 | 1.950 | 4.447 | 100 | 0.997 | 1.691 | 0.567 | 1.000 |
| Rothia dentocariosa | 93 | 1.136 | 2.277 | 100 | 0.347 | 0.342 | 0.652 | 0.775 |
| Streptococcus sanguinis | 100 | 5.920 | 5.214 | 100 | 2.753 | 3.034 | 0.045 | 1.000 |
| Veillonella dispar | 100 | 1.528 | 2.201 | 100 | 1.092 | 1.218 | 1.000 | 1.000 |
p<0.001. Mann–Whitney U test with Benjamini–Hochberg correction (false discovery rate: 10%) of the multiple comparisons. Bacterial species are shown with an average relative proportion ≥1% in the microbiome of both groups.
Profile of oral biofilm bacterial species according to type of dentition. The bacterial species with greatest relative abundance are shown. The area of each colour corresponds to the average relative abundance (average relative proportion) of each bacterial species in relation to the oral biofilm microbiome.
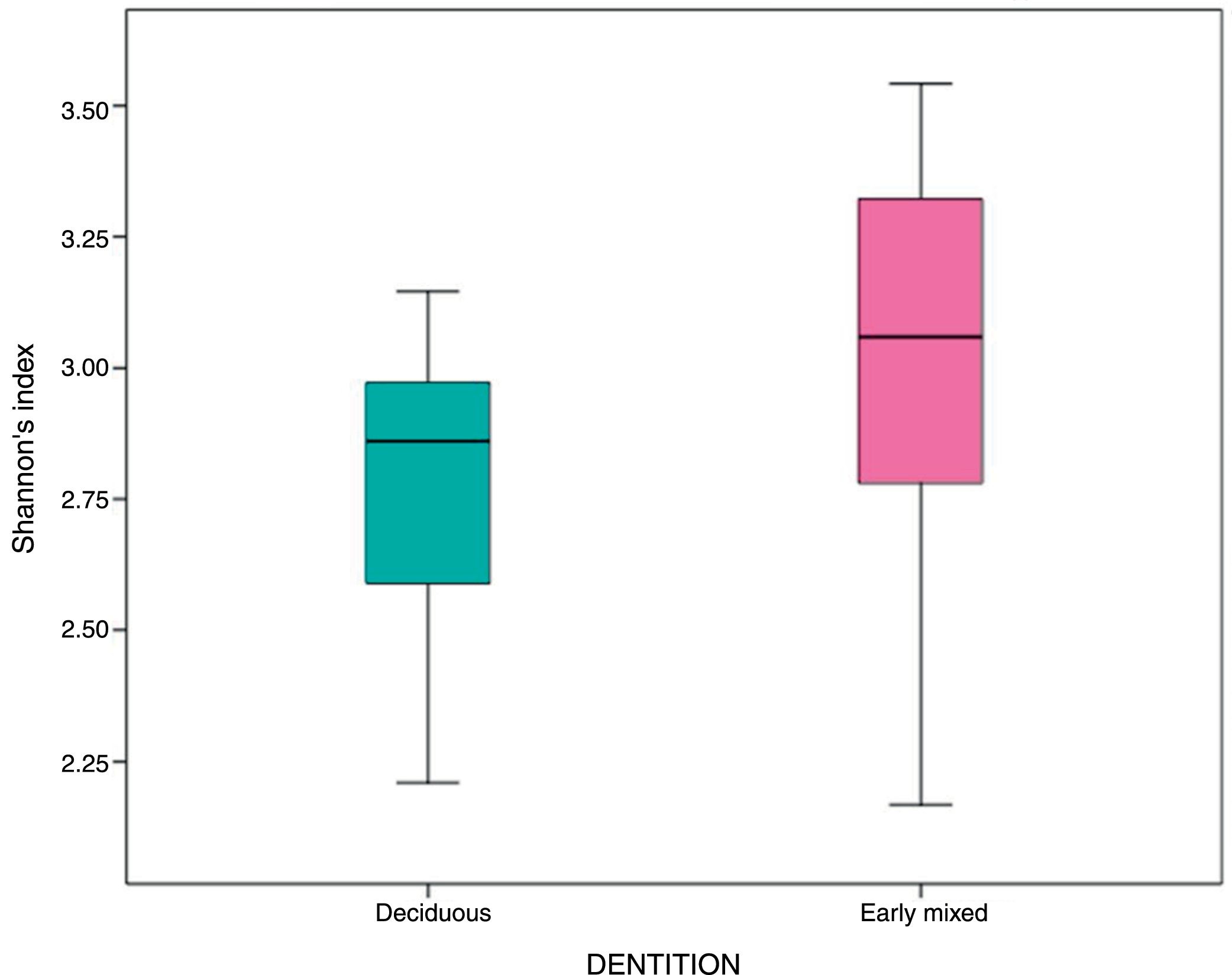
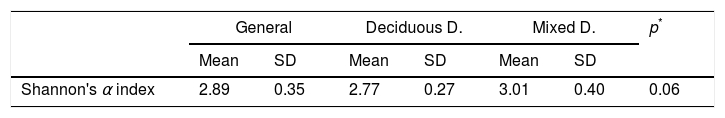
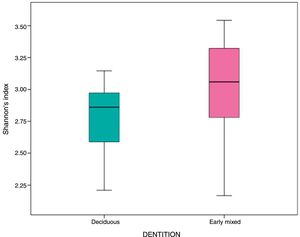
In the oral biofilm samples analysed, a bacterial species richness represented by Shannon's α diversity index of 2.89 (SD: 0.35) was found. This microbiological richness was consistent in the two study groups, with no statistically significant difference (p=0.06) (Table 3, Fig. 2).
Shannon's α microbiological diversity index.
| General | Deciduous D. | Mixed D. | p* | ||||
|---|---|---|---|---|---|---|---|
| Mean | SD | Mean | SD | Mean | SD | ||
| Shannon's α index | 2.89 | 0.35 | 2.77 | 0.27 | 3.01 | 0.40 | 0.06 |
Large quantities of bacteria can be found in the teeth, which form the bacterial biofilm. Some have been implicated in oral diseases. Research aiming to increase our understanding of the composition of the supragingival biofilm microbiome is important. Kamma et al. investigated the composition of the microbiota of the subgingival plaque of the different groups of teeth in children with mixed dentition. They reported having identified different groups of microorganisms, both in deciduous and permanent teeth, and they isolated 45 different microbial species.15 Xin et al. identified the oral microbial diversity of healthy Chinese children. They took samples of dental plaque from 10 children and in the results reported a total of 41 genera and 112 bacterial species.16 Shi et al. conducted a study in which they investigated the microbial profiles of the supragingival plaque of deciduous and permanent teeth in healthy children with mixed dentition and detected a total of 21 classes, 38 orders, 66 families and 74 genera,17 which confirms the microbial variety among deciduous and permanent teeth. This is consistent with our study, which describes the bacterial profile of the dental biofilm in children with deciduous and early mixed dentition; 44 genera and 360 bacterial species were found in the oral biofilm analysed, showing the considerable differences in the microbial diversity between mixed and deciduous dentition. These differences could be related to physical, chemical and environmental factors.
With regard to the time factor, Facal et al. studied the topography of caries in deciduous molars and their relationship to chronological age. They found that the physical form of the crown of a deciduous tooth varies from that of a permanent tooth: deciduous teeth have a small crown, with shallow trenches and grooves; permanent teeth have a larger crown with deeper pits and furrows, which creates areas of retention.18 In our study, the deciduous teeth had had greater opportunity for the development and maturation of the supragingival biofilm as they had been exposed to the environment in the mouth for longer (6.5 years) than the permanent teeth (5 years). The nutritional factor is important for the environment in the mouth: the deciduous dentition has been exposed to chemical substances involved in nutrition such as artificial milk and also both semi-solid and solid foods, while the mixed dentition has been exposed to solid foods, but not to artificial milk. All these nutritional factors could be associated with the microbiological diversity found.
With respect to bacterial genera, Shi et al. studied the microbial profiles of the supragingival biofilm of deciduous and permanent teeth and reported that there are differences in microbial diversity. From the samples they obtained, differences were found in permanent teeth in 25.2%; when compared with deciduous teeth, in 20.9%. The difference in the microbial community at the genus level was mainly due to Actinomyces for permanent dentition and Treponema for deciduous dentition,17 in line with our study, in which microbial diversity was found in the study samples from early mixed and deciduous dentition. However, there is a discrepancy in the difference in the bacterial community at genus level as the bacteria genera with greater relative abundance which differed from the microbial community in the early mixed dentition were Leptotrichia probe 3, TM7 genus and Neisseria genus probe 2, whereas in deciduous dentition, they were Actinomyces genus probe 3 and Fusobacterium probe 4.
Xu et al. studied the microbiome of the bacterial biofilm in children with deciduous teeth with and without caries. The most common genera in the group of patients without caries were Leptotrichia, Streptococcus, Actinomyces, Prevotella, Porphyromonas Neisseria and Veillonella19; some of which, i.e. Streptococcus probe 4, Streptococcus probe 1, Actinomyces probe 3, Veillonella probe 2 and Porphyromonas probe 2, are consistent with our study. However, they did not find the genus Fusobacterium probe 4.
ConclusionThe genera with greatest relative abundance in the biofilm samples from children with deciduous dentition were Streptococcus probe 4, Streptococcus probe 1, Actinomyces probe 3, Veillonella probe 2, Fusobacterium probe 4 and Porphyromonas probe 2; while in children with early mixed dentition, we found Streptococcus probe 4, Streptococcus probe 1, Leptotrichia probe 3, TM7 genus, Porphyromonas probe 2, Veillonella probe 2 and Neisseria genus. As far as bacterial species were concerned, those with greatest relative abundance in the samples from children with deciduous dentition were Streptococcus sanguinis, Rothia aeria, Gemella haemolysans, Gemella morbillorum, Veillonella dispar and Actinobaculum sp. HOT 183. In children with early mixed dentition, we found Streptococcus sanguinis, Leptotrichia sp. HOT 417, Leptotrichia sp. HOT 498, Gemella haemolysans, Granulicatella adiacens and Ottowia sp. HOT 894.
The bacterial profile of the supragingival dental biofilm in children with deciduous and early mixed dentition shows little microbiological diversity, either in presence and relative abundance or in terms of genus and bacterial species, with none of the results being statistically significant.
RecommendationsAs a recommendation for future studies, we would suggest increasing the sample size.
FundingThis study was funded by Colciencias (contract 673-2014), code No. 141965741160, and was conducted by Corporación Universitaria Rafael Núñez.
Conflicts of interestThe authors declare that they have no conflicts of interest.
The authors would like to thank Colciencias for funding this project with contract No. 673 of 2014, and the participants for taking part in the study.
Please cite this article as: Harris-Ricardo J, Fang L, Herrera-Herrera A, Fortich-Mesa N, Olier-Castillo D, Cavanzo-Rojas D, et al. Perfil bacteriano del biofilm dental supragingival en niños con dentición temporal y mixta temprana utilizando la técnica de secuenciación de próxima generación (HOMINGS). Enferm Infecc Microbiol Clin. 2019;37:448–453.