We aimed to determine the impact of utilizing a rapid panel test of respiratory viral and atypical bacteria (FilmArray® Respiratory Panel, FA RP) on etiological diagnosis of acute lower respiratory infection (ALRI) and antimicrobial stewardship in critical care pediatric patients.
MethodsProspective cohort study of patients aged<18 years with clinical diagnosis of ALRI that were admitted to the Pediatric Intensive Care Unit (PICU) of Hospital Sant Joan de Deu (Barcelona, Spain) during December 2015–February 2017. Patients were diagnosed by FA RP and by a bundle of routine microbiological assays.
ResultsALRI viral and bacterial etiology was confirmed by a composite reference standard of routine microbiological assays in 72 (55.4%) and 15 (11.5%) respiratory samples, respectively, that were collected from 130 children (median age, 3.5 months, IQR 1.1–14.8 months; 54.6% male). Comparatively, FA RP use increased etiological confirmation of ALRI in up to 123 (94.6%) samples (p<0.001) but only determined a bacterial origin in 2 (1.5%). Availability of diagnostic results before patient discharge from the PICU rose from 65.4 to 38.5% (p<0.001). Use of the new panel test directly influenced antimicrobial stewardship in 11 (8.4%) episodes, leading to discontinuation of antiviral drugs (n=5), administration of targeted antibiotics (n=3), antiviral therapy start (n=2) and both targeted antibiotic administration and discontinuation of antiviral drugs (n=1).
ConclusionFA RP contributed to improve etiological diagnosis of ALRI in a timely manner while enhancing a more rational use of antimicrobial drugs in critical care pediatric patients.
Nuestro objetivo fue determinar el impacto de la utilización de una prueba rápida de detección múltiple de virus y bacterias atípicas respiratorias (FilmArray® Respiratory Panel [FA RP]) en el diagnóstico etiológico de la infección respiratoria aguda de vías bajas (IRAVB) y en la administración de antimicrobianos en pacientes críticos pediátricos.
MétodosEstudio de una cohorte prospectiva de pacientes <18 años con diagnóstico clínico de IRAVB que ingresaron en la Unidad de Cuidados Intensivos Pediátricos (UCIP) del Hospital Sant Joan de Deu, Barcelona, España, durante diciembre de 2015-febrero de 2017. Los pacientes fueron diagnosticados por FA RP y por un grupo de pruebas microbiológicas de rutina.
ResultadosLas pruebas microbiológicas de rutina confirmaron la etiología viral y bacteriana de la IRAVB en 72 (55,4%) y 15 (11,5%) muestras respiratorias, respectivamente, obtenidas de 130 niños (edad mediana: 3,5 meses; rango intercuartil: 1,1-14,8 meses; 54,6% varones). Comparativamente, el uso de FA RP aumentó la confirmación etiológica de la IRAVB en hasta 123 (94,6%) muestras (p<0,001), pero solo determinó un origen bacteriano en 2 (1,5%). La disponibilidad de resultados diagnósticos antes del alta del paciente de la UCIP aumentó del 38,5 al 65,4% (p<0,001). El uso de la nueva prueba de detección múltiple influyó directamente en la administración de antimicrobianos en 11 (8,4%) episodios, orientando la interrupción de tratamientos antivirales (n=5), la administración de antibióticos dirigidos (n=3), el inicio de terapias antivirales (n=2) y la administración dirigida de antibióticos e interrupción simultánea de tratamiento antiviral (n=1).
ConclusiónFA RP contribuyó a mejorar y agilizar el diagnóstico etiológico de la IRAVB, facilitando un uso más racional de antimicrobianos en pacientes críticos pediátricos.
Acute lower respiratory infection (ALRI) remains the most important global public health problem amongchildren.1–3 ALRI was estimated to cause14.9 million episodes that resulted in pediatric hospital admissions worldwide in 20104 and around 650,000 deaths of young children in 2016.5 The disease is caused by a large and heterogeneous group of infections including bacterial, viral, and other etiologies. Although incidence of viral ALRI is larger at early ages,6,7 pathogenic respiratory bacteria produce higher morbidity and mortality rates globally, especially among older children.5 Occurrence of mixed ALRI is also common during childhood.8–10
Distinguishing the etiology of ALRI becomes challenging, since derived signs and symptoms are often unspecific. A gold standard for etiological diagnosis of ALRI has not yet been developed.11 Conventional microbiological diagnostic methods such as bacterial culture, targeted polymerase-chain reaction (PCR) assays and rapid viral antigen tests have limitations in comprehensiveness, accuracy, and/or timeliness of results to guide clinical decisions, even expanding the arsenal of diagnostic tools with chest radiographies and acute-phase reactant measurements.12,13
Treatment of children with viral ALRI appears to be an area where extensive misuse of antibiotics could be reduced.14 In the last years, the introduction of fast molecular assays for multiple identification of respiratory viruses and bacteria has offered clinicians the potential to identify the viral origin of ALRI that might otherwise be considered to have a bacterial etiology and thus be treated with antibiotics. The FilmArray® Respiratory Panel (BioFire Diagnostics Inc., US), hereafter FA RP, is a qualitative reverse transcriptase PCR panel assay that targets adenovirus (AdV), coronavirus (CoV) types 229E/NL63/OC43/HKU1, influenza A virus (IFV-A) including differentiation of subtypes H1/H1N1-2009/H3, influenza B virus (IFV-B), human metapneumovirus (HMPV), parainfluenza virus (PIV) types 1/2/3/4, respiratory syncytial virus (RSV) types A/B, rhinovirus/enterovirus (RV/EV), Bordetella pertussis, Chlamydia pneumoniae, and Mycoplasma pneumoniae in a single respiratory sample. FA RP integrates sample preparation, DNA amplification and detection into an automated process with only 2min of hands-on time and 1 hour of instrumentation time.15
The main aim of our study was to evaluate to which extent the utilization of FA RP improved etiological diagnosis of ALRI and subsequent antimicrobial prescribing practices in critical care pediatric patients, compared to a bundle of routine microbiological diagnostic assays.
Materials and methodsStudy design and settingA prospective single-center cohort study of pediatric patients with clinical diagnosis of ALRI admitted to the Pediatric Intensive Care Unit (PICU) of Hospital Sant Joan de Deu (Barcelona, Spain) was conducted during the period December 2015-February 2017. All patients with clinical suspicion of ALRI within such period were initially considered for inclusion in the study. Participant inclusion criteria were: (1) age<18 years; (2) clinical presentation compatible with acute respiratory disease (cough, difficult breathing, tachypnea) and/or signs and symptoms of infection (reported or documented fever>37.3°C or looking/feeling unwell); and (3) informed consent to participate obtained from parents or guardians. Patients were excluded from the study if they had been hospitalized in the previous 14 days before the current episode or infection onset occurred 48h after the date of PICU admission. The study setting is a 318-bedsize reference university hospital that attends a reference population of approximately 300,000 children.
All patients were diagnosed by FA RP as well as by a bundle of routine microbiological assays. Primary endpoints were FA RP diagnostic performance compared to a composite reference standard of the bundle of routine microbiological diagnostic assays, antimicrobial prescription changes made as a consequence of FA RP results, and days of antimicrobials saved that could be attributed to the use of the panel test. Secondary outcomes were patient baseline characteristics and length of PICU stay. Clinical data, data of antimicrobial use and laboratory diagnostic results and timeliness were retrieved from the Electronic medical records and the Pharmacy and Laboratory Information systems of the study setting. The study was approved by the Ethics Committee of the site and informed consent was obtained from parents or guardians of participants for patient enrolment.
DefinitionsViral etiology of ALRI was microbiologically confirmed by detection of respiratory virus genetic material by FA RP or a single viral PCR assay or by antigen detection of RSV or IFV in nasopharyngeal aspirates, bronchoalveolar lavages or endotracheal aspirates. A bacterial culture negative result was considered to be suggestive of a viral etiology. Bacterial ALRI etiology was microbiologically confirmed by observation of pathogenic bacterial growth by culture or bacterial nucleic acid detection in blood, bronchoalveolar lavages or endotracheal aspirates.
Sample collection and microbiological methodsFresh respiratory samples were collected from patients and processed on demand by FA RP and routine tests according to standard operational procedures and to the manufacturers’ instructions in the clinical laboratory of the setting. Requests for routine diagnostic tests were made at the discretion of clinicians based on patient presentation and history and, in the case of single PCRs, were jointly agreed with the microbiologists of the clinical laboratory on a case-per-case basis. PCR testing timetable in the clinical laboratory of the study setting was Monday to Friday from 7.00am to 8.00pm.
Statistical analysisDescriptive variables were analyzed using means and standard deviations (SD), medians and interquartile ranges (IQR), or frequencies and percentages, as appropriate. Time to result by FA RP was calculated as the time elapse since sample receipt in the clinical laboratory of the setting until result registration in the Laboratory Information system. Significance of the difference between the proportion of antimicrobial prescriptions at baseline and after availability of FA RP results was determined by the Chi-square or the Fisher exact test. All statistical analyses were performed using Stata v.15.1 software (Stata Corp., USA).
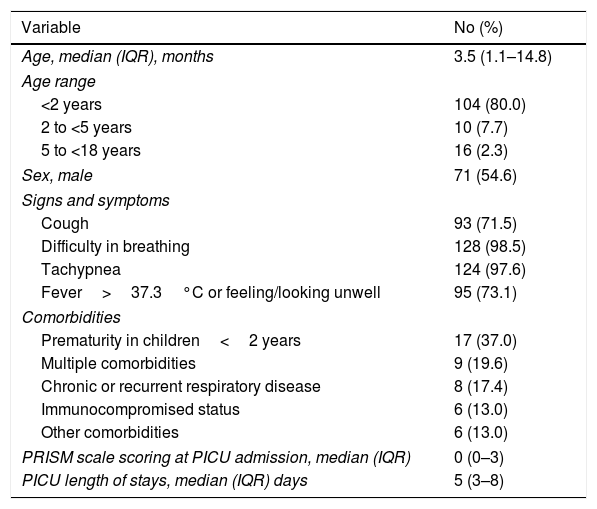
ResultsBaseline patient characteristics and PICU length of stayOne-hundred and seventy-four patients were screened for inclusion in the study. Of them, 44 (25.3%) were discarded because informed consent could not be obtained (n=36) or due to previous recent hospitalization or infection onset after 48hours since PICU admission (n=8). A total of 130 patients were finally selected for the study. Median age of participants was 3.5 (IQR, 1.1–14.8) months and 71 (54.6%) were male. Forty-six (35.4%) participants presented co-morbidities, being prematurity in infants<2 years of age the most predominant co-morbid condition (n=17, 37.0%). Baseline characteristics of participants are described in Table 1. Median length of stay in PICU was 5 days (IQR, 3–8 days).
Baseline and follow-up cohort characteristics.
| Variable | No (%) |
|---|---|
| Age, median (IQR), months | 3.5 (1.1–14.8) |
| Age range | |
| <2 years | 104 (80.0) |
| 2 to <5 years | 10 (7.7) |
| 5 to <18 years | 16 (2.3) |
| Sex, male | 71 (54.6) |
| Signs and symptoms | |
| Cough | 93 (71.5) |
| Difficulty in breathing | 128 (98.5) |
| Tachypnea | 124 (97.6) |
| Fever>37.3°C or feeling/looking unwell | 95 (73.1) |
| Comorbidities | |
| Prematurity in children<2 years | 17 (37.0) |
| Multiple comorbidities | 9 (19.6) |
| Chronic or recurrent respiratory disease | 8 (17.4) |
| Immunocompromised status | 6 (13.0) |
| Other comorbidities | 6 (13.0) |
| PRISM scale scoring at PICU admission, median (IQR) | 0 (0–3) |
| PICU length of stays, median (IQR) days | 5 (3–8) |
Values expressed as No. (%), unless otherwise stated.
Abbreviations: IQR, interquartile range; PICU, Pediatric Intensive Care Unit; PRISM, Pediatric Risk of Mortality.
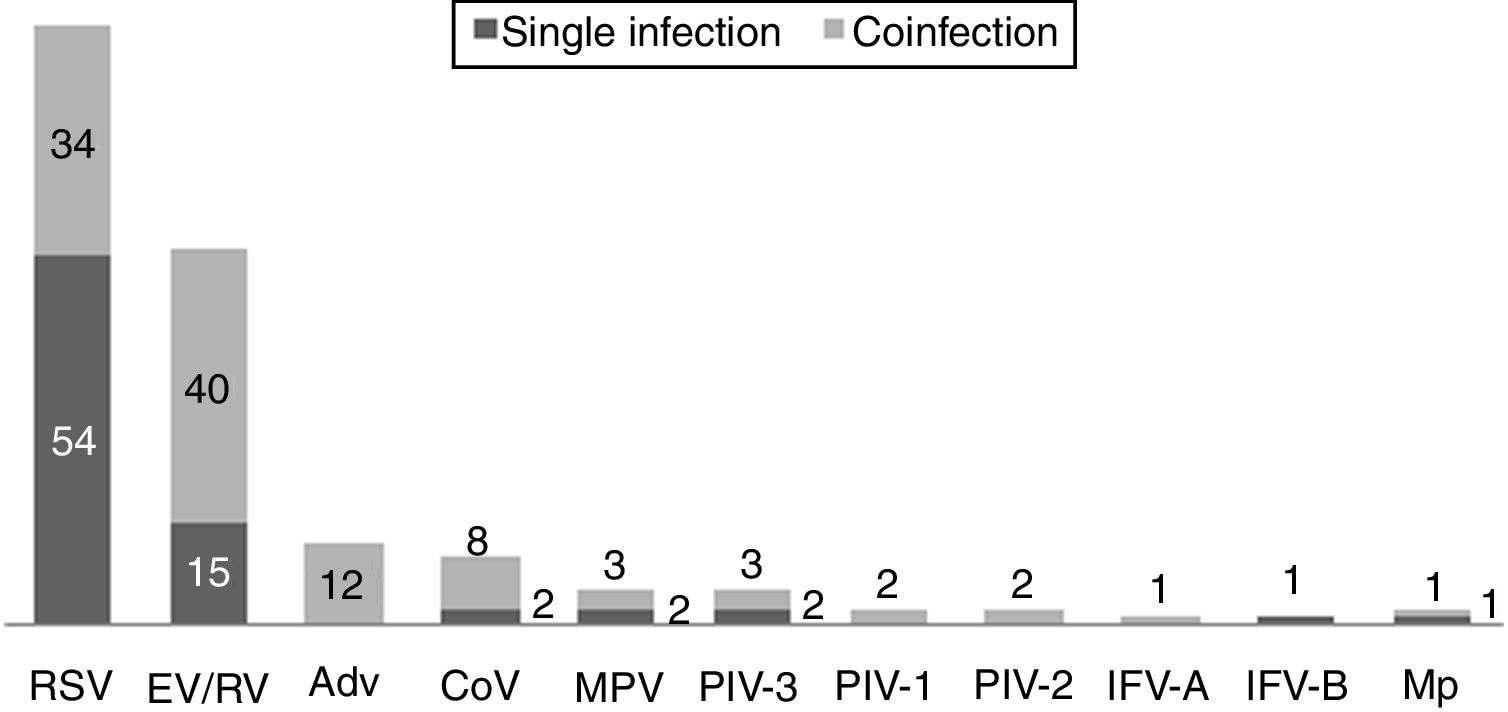
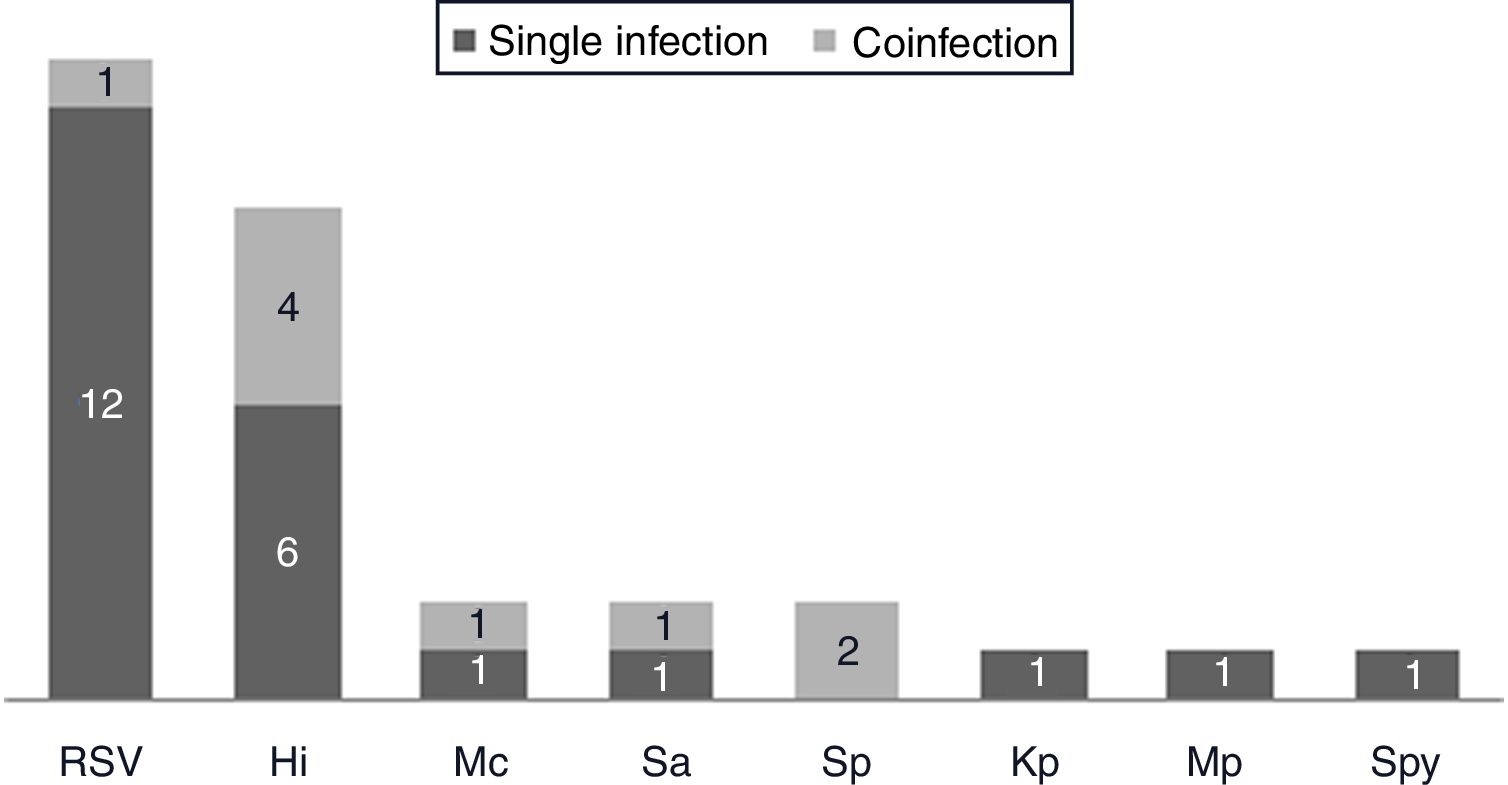
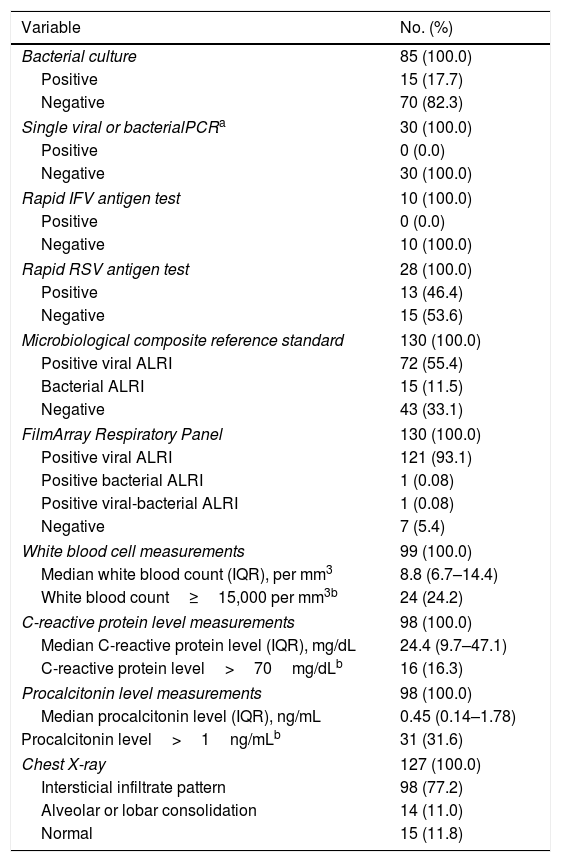
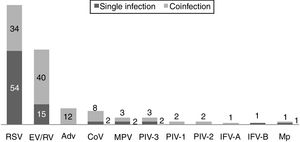
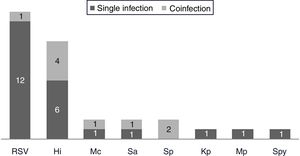
One hundred and twenty-two (93.9%) nasopharyngeal aspirates, 5 (3.9%) tracheal aspirates, and 3 (2.3%) bronchoalveolar lavages were collected and tested by FA RP. The panel test detected 123 (94.6%) positive specimens: 121 (93.1%) contained viruses and 2 (1.5%) contained atypical bacteria, specifically Mycoplasma pneumoniae. RSV was the most prevalent species detected in samples (n=88), followed by EV/RV (n=55). Viral co-detections were common (n=43, 35.0%) while identification of bacterial-viral co-detections was infrequent (n=1, 0.8%). Targets most commonly involved in co-detections were EV/RV (n=40) and RSV (n=34). The composite reference standard identified 87 (66.9%) positive samples, being 72 (55.4%) infected by viruses and 15 (11.5%) by bacteria. Most prevalent respiratory pathogens routinely identified were RSV (13 specimens that tested positive by rapid antigen tests) and Haemophilus influenzae (10 specimens that tested positive by bacterial culture). Four bacterial coinfections and 1 bacterial-viral coinfection were observed by the bundle of routine diagnostic assays, mostly involving Haemophilus influenzae (n=5) and Streptococcus pneumoniae (n=2). Overall, FA RP significantly increased diagnostic yield of routine diagnostic assays from 66.9 to 93.9% (p<0.001). This increase was due to a comparatively much higher viral detection rate of FA RP (93.1 vs.55.4%) but not to the capability of the panel test to detect pathogenic bacteria (1.5 vs. 11.5%). Figs. 1 and 2 depict the distribution of respiratory pathogens in the study cohort. Table 2 details results of laboratory and imaging diagnostic tests. Table 3 describes pathogen combinations identified in co-infected samples.
Respiratory pathogen distribution by FilmArray Respiratory Panel. Abbreviations: RSV, respiratory syncytial virus; RV, rhinovirus; AdV, adenovirus; CoV, coronavirus; EV, enterovirus; MPV, metapneumovirus; PIV-1/2/3, parainfluenza virus 1/2/3; IFV-A/B, influenza virus A/B; Mp, Mycoplasma pneumoniae.
Respiratory pathogen distribution by other microbiological tests. Abbreviations: RSV, respiratory syncytial virus; Hi, Haemophilus influenzae; Mc, Moraxella catarrhalis; Sa, Staphlylococcus aureus; Sp, Streptococcus pneumoniae; Kp, Klebsiella pneumoniae; Mp, Mycoplasma pneumoniae; Spy, Streptococcus pyogenes.
Laboratory and imaging diagnostic test results.
| Variable | No. (%) |
|---|---|
| Bacterial culture | 85 (100.0) |
| Positive | 15 (17.7) |
| Negative | 70 (82.3) |
| Single viral or bacterialPCRa | 30 (100.0) |
| Positive | 0 (0.0) |
| Negative | 30 (100.0) |
| Rapid IFV antigen test | 10 (100.0) |
| Positive | 0 (0.0) |
| Negative | 10 (100.0) |
| Rapid RSV antigen test | 28 (100.0) |
| Positive | 13 (46.4) |
| Negative | 15 (53.6) |
| Microbiological composite reference standard | 130 (100.0) |
| Positive viral ALRI | 72 (55.4) |
| Bacterial ALRI | 15 (11.5) |
| Negative | 43 (33.1) |
| FilmArray Respiratory Panel | 130 (100.0) |
| Positive viral ALRI | 121 (93.1) |
| Positive bacterial ALRI | 1 (0.08) |
| Positive viral-bacterial ALRI | 1 (0.08) |
| Negative | 7 (5.4) |
| White blood cell measurements | 99 (100.0) |
| Median white blood count (IQR), per mm3 | 8.8 (6.7–14.4) |
| White blood count≥15,000 per mm3b | 24 (24.2) |
| C-reactive protein level measurements | 98 (100.0) |
| Median C-reactive protein level (IQR), mg/dL | 24.4 (9.7–47.1) |
| C-reactive protein level>70mg/dLb | 16 (16.3) |
| Procalcitonin level measurements | 98 (100.0) |
| Median procalcitonin level (IQR), ng/mL | 0.45 (0.14–1.78) |
| Procalcitonin level>1ng/mLb | 31 (31.6) |
| Chest X-ray | 127 (100.0) |
| Intersticial infiltrate pattern | 98 (77.2) |
| Alveolar or lobar consolidation | 14 (11.0) |
| Normal | 15 (11.8) |
Values expressed as No. (%), unless otherwise stated.
Distribution of respiratory co-detections.
| Target | No (%) |
|---|---|
| Co-detections by FilmArray Respiratory Panel | 44 (35.8) |
| AdV+EV/RVa+MPV | 1 (0.8) |
| AdV+EV/RVa+PIV-1 | 1 (0.8) |
| AdV+EV/RVa+RSV | 1 (0.8) |
| CoV+EV/RVa+RSV | 1 (0.8) |
| CoV+PIV-3+RSV+EV/RVa | 1 (0.8) |
| EV/RVa+PIV-2+RSV | 1 (0.8) |
| EV/RVa+PIV-3+RSV | 1 (0.8) |
| AdV+RSV+EV/RVa | 5 (4.1) |
| AdV+CoV+EV/RVa | 1 (0.8) |
| PIV-3+RSV+EV/RVa | 1 (0.8) |
| RSV+EV/RVa | 18 (14.6) |
| CoV+RSV | 4 (3.3) |
| AdV+EV/RVa | 2 (1.6) |
| AdV+PIV-1 | 1 (0.8) |
| CoV+MPV | 1 (0.8) |
| IFV-A+EV/RVa | 1 (0.8) |
| MPV+EV/RVa | 1 (0.8) |
| PIV-2+RSV | 1 (0.8) |
| EV/RVa+Mp | 1 (0.8) |
| Co-infections by other microbiologicaltestsb | 5 (4.1) |
| Hi+Sp | 2 (1.8) |
| Hi+Mc | 1 (0.8) |
| Hi+Sa | 1 (0.8) |
| Hi+RSV | 1 (0.8) |
Abbreviations: AdV, adenovirus; BoV, bocavirus; CoV, coronavirus; EV/RV, enterovirus/rhinovirus; IFV-A/B, influenza virus A/B; MPV, metapneumovirus; PIV-1/2/3/4, parainfluenza virus; RSV, respiratory syncytial virus; Hi, Haemophilus influenzae; Mc, Moraxella catarrhalis; Mp, Mycoplasma pneumoniae; Sa, Staphylococcus aureus; Sp, Streptococcus pneumoniae; Kp, Klebsiella pneumoniae; Mp, Mycoplasma pneumoniae; Spy, Streptococcus pyogenes.
Median time to result by FA RP was 2.9h (IQR, 2.2–5.0h) for samples received and processed within PCR testing timetable. Panel test results were available for 85 (65.4%) patients before discharge from the PICU whereas results of routine microbiological assays were only available for 50 (38.5%, p<0.001) patients. The low proportion of ALRI etiologies confirmed by routine microbiological methods during patient stay in the PICU was due to the prolonged time to results of bacterial culture (≥48h).
Contribution of FA RP to antimicrobial stewardshipDiverse strategies of antibiotic treatment were implemented after delivery of FA RP results: baseline antibiotics were maintained in 74 (57.0%) children, 28 (21.5%) children remained off antibiotics, and 28 (21.5%) had baseline antibiotic prescriptions modified. Antibiotic treatment changes resulted in discontinuation of antibiotherapies (n=20), implementation of more targeted antibiotic therapies (n=6), and antibiotic start (n=2). FA RP directly orientated changes to a more targeted antibiotic therapy in 4 patients: antibiotics were de-escalated after FA RP negative results for Mycoplasma pneumoniae in 3 and escalated in 1 after FA RP positive results for that bacterium. Influence of FA RP use on the remainder 24 antibiotic changes could not be determined, since those changes were also driven by a sum of factors including patient evolution, diagnostic results of routine microbiological assays, and prognostic indications of acute-phase reactants.
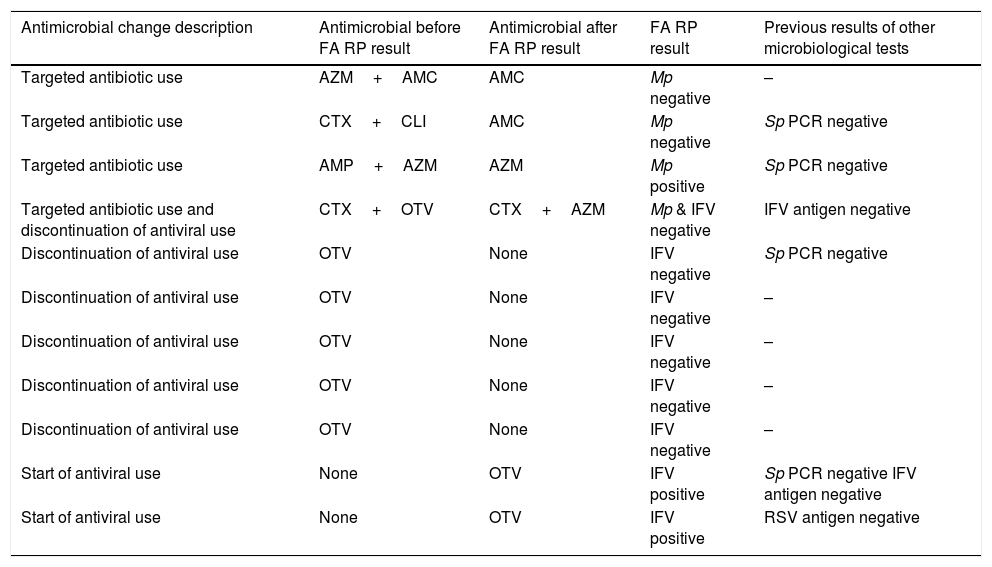
The effect of FA RP use on administration of antiviral drugs was observed in 8 (6.2%) children: antivirals were discontinued in 6 of them after FA RP negative results for IFV and were started in 2 after FA RP positive results for IFV (otherwise undetected by routine microbiological assays). Discontinuation of antiviral use saved 4 days per episode with antiviral treatment, considering 5 days as a standard duration of antiviral use in children. In total, antimicrobial stewardship changes solely due to FA RP utilization were implemented in 11 (8.5%) patients of the cohort, as detailed in Table 4.
Antimicrobial stewardship changes orientated by FilmArray Respiratory Panel results.
| Antimicrobial change description | Antimicrobial before FA RP result | Antimicrobial after FA RP result | FA RP result | Previous results of other microbiological tests |
|---|---|---|---|---|
| Targeted antibiotic use | AZM+AMC | AMC | Mp negative | – |
| Targeted antibiotic use | CTX+CLI | AMC | Mp negative | Sp PCR negative |
| Targeted antibiotic use | AMP+AZM | AZM | Mp positive | Sp PCR negative |
| Targeted antibiotic use and discontinuation of antiviral use | CTX+OTV | CTX+AZM | Mp & IFV negative | IFV antigen negative |
| Discontinuation of antiviral use | OTV | None | IFV negative | Sp PCR negative |
| Discontinuation of antiviral use | OTV | None | IFV negative | – |
| Discontinuation of antiviral use | OTV | None | IFV negative | – |
| Discontinuation of antiviral use | OTV | None | IFV negative | – |
| Discontinuation of antiviral use | OTV | None | IFV negative | – |
| Start of antiviral use | None | OTV | IFV positive | Sp PCR negative IFV antigen negative |
| Start of antiviral use | None | OTV | IFV positive | RSV antigen negative |
Abbreviations: FA RP, FilmArrayRespiratory panel; PCR, polymerase-chainreaction; Mp, Mycoplasmapneumoniae; Sp, Streptococcus pneumoniae; IFV, influenza virus; RSV, respiratory syncytial virus; AMC, amoxicillin-clavulaninacid; AMP, ampicillin; AZM, azythromycin; CLI, clarithromycin; CTX, cefotaxime; OTV, oseltamivir.
Our study showed an increase in diagnostic yield, timeliness of results and judicious use of antimicrobials as a consequence of the implementation of FA RP for diagnosing ALRI, in comparison with conventional microbiological tests. To the best of our knowledge, these findings had not been previously reported in cohorts of critical care pediatric patients, a specific study group characterized by a noticeable proportion of bacterial ALRI and mixed ALRI etiologies.
An increase in diagnostic yield attributable to FA was observed in precedent studies that compared accuracy of the panel test with that of other viral panel tests16 and batched PCR assays.17 Nonetheless, previous diagnostic accuracy studies were mostly focused on adult cohorts or groups. On the other hand, FA RP median time to results observed in our study (2.9h) was consistent with outcomes reported in previous studies reporting a mean time of 3.1h18 or median times of 1.4h16,19 and 2.3h20 for FA RP testing.
Previous literature on the potential linkage of rapid respiratory panel testing with optimized antibiotic use shows discordant results. A study in pediatric and adult patients with uncomplicated ARI admitted to an Emergency Department during the influenza epidemic season described a decrease in antibiotic use of half a day after shifting from a set of multiplex and singleplex commercial PCR assays to FA RP.17 In contrast, a randomized controlled trial of adults presenting with ARI to the Emergency Department or acute medical unit of a large hospital over two winter seasons and tested either by FA RP or laboratory PCR tests reported that mean duration of antibiotics was similar in both groups.20 Similarly, a quasi-randomized study in adults hospitalized with upper respiratory infection or influenza-like illness, with or without lower respiratory infection, found no evidence of reductions in antibiotic utilization as a consequence of FA RP testing, compared to equivalent outcomes when testing was performed by an array of laboratory-developed multiplex and singleplex PCR assays.21 Moreover, an observational study conducted in a hospital that switched from a respiratory viral panel to FA RP for diagnosing adult patients with respiratory viral illnesses did not observe statistically significant differences in antibiotic use after the change in the diagnostic strategy, yet time to results decreased markedly from 24 to 12h.22
We speculate that we found a minor decline in antibiotic use directly associated to FA RP implementation because the presence of a respiratory virus detected by a panel test that covers an extensive set of viral targets but only certain atypical bacterial targets does not rule out a potential bacterial coinfection, particularly in a PICU environment where patients are highly vulnerable. The marked difference between proportions of bacterial ALRI etiologies confirmed by FA RP and the composite reference standard (1.5 vs. 11.5%) supports our hypothesis. Withdrawing or postponing antibiotic administration in the PICU before availability of negative bacterial culture results in the absence of a comprehensive bacterial and viral respiratory panel test appears unlikely, given the high risk of concomitant or secondary bacterial ALRI in severely ill children. Results suggest the need of combining FA RP with bacterium-targeted microbiological assays on appropriate pediatric respiratory specimens (bronchoalveolar lavage or endotracheal aspirates) for comprehensive etiological diagnosis of pediatric severe ALRI. In this regard, a general recommendation has been made for rapid molecular diagnostic tests to incorporate testing for relevant bacterial pathogens in addition to viral targets in order to limit antibacterial therapy.23 It is also to be noted that the manufacturer of FA RP has recently launched a pneumonia panel test that includes 18 bacterial pathogens, some of which are highly prevalent in bacterial ALRI.24
Interestingly, we observed that detection or not detection of Mycoplasma pneumoniae by FA RP was a factor leading to a change to a more targeted antibiotic therapy, either for escalation or de-escalation, in line with recommendations of clinical practice guidelines for treatment of community-acquired pneumonia in children.25 In a similar way, detection or not detection of IFV by the panel test guided discontinuation or start of antiviral treatment, as recommended by guidelines. It is worthwhile to highlight that IFV-positive samples by FA RP were not bacterial-positive by other microbiological assays and patients sampled did not show radiological findings or acute-phase reactant measurements that could suggest bacterial co-infection and thus remained off antibiotics, also in accordance with guidelines.
This study presents some limitations. First, it was a single-center study with a relatively low sample size, although the cohort was adequately characterized and monitored to infer consistent conclusions. Second, requests for routine tests were made at the discretion of clinicians. This aspect might induce a certain bias depending on individual preferences to order certain microbiological assays and not others. However, in our view the study reflects the real situation in a hospital environment where some variability in the selection of the best diagnostic strategy may exist among clinicians. Third, outcomes were obtained using an observational cohort design. Further analysis adopting experimental randomized designs would increase external validity of results.
In conclusion, diagnostic performance of FA RP improved accuracy and timeliness of ALRI etiological diagnosis in critical care pediatric patients, in comparison to routine microbiological tests. Implementation of the panel test enhanced a more rational use of antiviral and antibiotic drugs in those patients. However, non negligible occurrence of bacterial ALRI in children suggests the need to combine the panel test with other bacterium-targeted microbiological assays for comprehensive etiological diagnosis of pediatric severe ALRI.
Conflict of interest statement: CMA reports research grants to her institution from Qiagen, Biofire Diagnostics, Pfizer, Roche Diagnostics, BioMérieux, Alere and Genomica SAU, and compensation fees from Qiagen, Roche Diagnostics and BioMérieux for scientific presentations in satellite symposiums; PB reports compensation fees from Roche Diagnostics for scientific presentations in satellite symposiums. The rest of authors report no conflict of interest.
Funding statement: the study was partially funded by Biofire Diagnostics and BioMérieux. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Conflict of interestCMA reports research grants to her institution from Qiagen, Biofire Diagnostics, Pfizer, Roche Diagnostics, BioMérieux, Alere and Genomica SAU, and compensation fees from Qiagen, Roche Diagnostics and BioMérieux for scientific presentations in satellite symposiums; PB reports compensation fees from Roche Diagnostics for scientific presentations in satellite symposiums. The rest of authors report no conflict of interest.