The aim was to evaluate the utility of a multiplex real-time PCR to detect Streptococcus pneumoniae lytA, plyA and psaA genes in pleural fluid (PF).
MethodsA collection of 81 PF samples was used. Sixty were considered positive for S. pneumoniae according to previous results (54 by an in-house lytA gene PCR and eight by universal rRNA PCR).
ResultsThe sensitivity for detection of the lytA, plyA and psaA genes by multiplex PCR was 100% (60/60), 98.3% (59/60) and 91.7% (55/60), respectively. The detection of all three genes was negative in 21 samples formerly confirmed as negative for S. pneumoniae (100% specificity) by the other procedures (9 by in-house lytA PCR and 12 by rRNA PCR).
ConclusionsThe use of this multiplex PCR may be a useful option to identify S. pneumoniae directly in PF samples.
El objetivo fue evaluar la utilidad de una técnica de PCR múltiple para detectar los genes lytA, plyA y psaA de Streptococcus pneumoniae en líquido pleural.
MétodosSe empleó una colección de 81 muestras de líquido pleural. Sesenta habían sido consideradas positivas para S. pneumoniae según resultados previos (54 por una prueba casera de PCR para el gen lytA y 8 por una PCR universal rRNA).
ResultadosLa sensibilidad de la técnica para la detección de los genes lytA, plyA y psaA fue respectivamente 100% (60/60), 98,3% (59/60) y 91,7% (55/60). La detección de los tres genes resultó negativa en 21 muestras negativas (especificidad 100%) por los otros procedimientos (9 por la prueba casera de PCR para lytA y 12 por la PCR rRNA).
ConclusionesEl uso de esta técnica de PCR múltiple puede ser una opción útil para la detección directa de S. pneumoniae en líquido pleural.
The gold standard for diagnosis of invasive pneumococcal disease (IPD) is microbiological culture from a usually sterile clinical sample. This procedure requires viable organisms for isolating. Culture-based methods have some advantages, including low cost and ability to provide strains for serotyping and antibiotic susceptibility testing.1 However, Streptococcus pneumoniae has the tendency to autolyse when reaching the stationary phase of growth1 resting sensitivity to the microbiological culture, mainly in some samples as pleural fluid (PF).2 Due to the low sensitivity of the culture, other alternatives for the diagnosis of IPD, including immunological detection assays and nucleic acid amplification, have been implemented.1,3,4 The direct immunological detection in clinical samples may be achieved by immunochromatographic testing of species specific pneumococcal soluble antigen5,6 and less frequently by serogroup/serotype antigen identification by polyclonal antisera prepared for latex agglutination.7,8 For the molecular identification of S. pneumoniae several targets have been employed, including lytA, plyA and psaA genes (that codifies respectively the main pneumococcal autolysin (LytA), the pneumolysin (Ply) and the lipoprotein component of a Mn2+ transporter codified (PsaA)).1,9 The aim of this study was to evaluate the utility of a new commercial multiplex real time polymerase chain reaction (RT-PCR) method (S. pneumoniae-VK; Vacunek, S.L. Derio, Bizkaia, Spain) for the detection of S. pneumoniae lytA, plyA and psaA genes in PF.
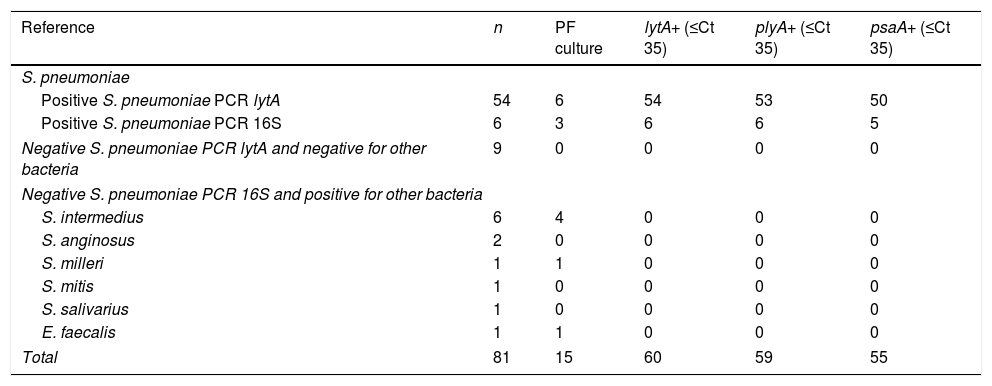
Materials and methodsA collection of 81 PF samples previously tested for clinical diagnosis purposes was employed (Table 1). All patients presented pleural effusion, in 66 the PF microbiological culture was negative and in 15 some bacterial strain was isolated (9 S. pneumoniae, 5 Streptococcus viridans [4 Streptococcus intermedius and 1 Streptococcus milleri] and 1 Enterococcus faecalis). Sixty samples (including the 9 culture positive) have been considered formerly positive for S. pneumoniae: 54 were positive to lytA gene by an in house PCR technique6,10 and 6 positive by universal PCR plus sequencing of the 16S ribosomal ribonucleic acid gene (16S rRNA PCR).11 Twenty one samples have been early considered negative for S. pneumoniae: 9 were negative to lytA gene by the in house PCR and 12 positive by the 16S PCR for other different species (6 for S. intermedius, 2 for Streptococcus anginosus, 1 for S. milleri, 1 for Streptococcus mitis, 1 for Streptococcus salivarius and 1 for E. faecalis). Streptococcus pneumoniae samples tested by each reference technique (lytA in house PCR or universal 16S rRNA PCR) were not assayed by the other.
Distribution of polymerase chain reaction (PCR) lytA, plyA and psaA results in the 81 pleural fluid (PF) studied samples.
| Reference | n | PF culture | lytA+ (≤Ct 35) | plyA+ (≤Ct 35) | psaA+ (≤Ct 35) |
|---|---|---|---|---|---|
| S. pneumoniae | |||||
| Positive S. pneumoniae PCR lytA | 54 | 6 | 54 | 53 | 50 |
| Positive S. pneumoniae PCR 16S | 6 | 3 | 6 | 6 | 5 |
| Negative S. pneumoniae PCR lytA and negative for other bacteria | 9 | 0 | 0 | 0 | 0 |
| Negative S. pneumoniae PCR 16S and positive for other bacteria | |||||
| S. intermedius | 6 | 4 | 0 | 0 | 0 |
| S. anginosus | 2 | 0 | 0 | 0 | 0 |
| S. milleri | 1 | 1 | 0 | 0 | 0 |
| S. mitis | 1 | 0 | 0 | 0 | 0 |
| S. salivarius | 1 | 0 | 0 | 0 | 0 |
| E. faecalis | 1 | 1 | 0 | 0 | 0 |
| Total | 81 | 15 | 60 | 59 | 55 |
Ct: threshold cycle.
PF samples were tested by S. pneumoniae-VK. This technique is a quadruplex RT-PCR that detects lytA, plyA and psaA genes9 and an internal control random sequence of 117 pair of bases not coinciding with any previously NCBI described one. The Ct was considered as the PCR cycle at which an increase in the fluorescence signal is detected initially. Positive lytA, plyA and psaA results were considered for Ct ≤35.
Ninety five per cent confidence intervals of proportions of categorical variables with two possible outcomes were calculated by the modified Wald method by using the ©2017 GraphPad Software.
ResultsTable 1 shows the distribution of PCR lytA, plyA and psaA results in the 81 PF studied samples. lytA, plyA and psaA gene detection were respectively positive in 60, 59 and 55 out of 60 S. pneumoniae formerly positive samples (giving values of sensitivity of 100% [IC95% 92.8–100] for lytA, 98.3% [IC95% 90.3 to >99.9] for plyA and 91.7% [IC95% 81.5–96.8] for psaA). The 59 samples plyA positive were also positive for lytA and the 55 samples positive for psaA were simultaneously positive for plyA and psaA. Inversely, lytA, plyA and psaA detection were negative in all the three genes in 21 out of 21 S. pneumoniae formerly (9 in house PCR lytA or 12 PCR 16S) negative samples (giving for lytA, plyA and psaA genes values of specificity of 100% [IC95% 81.8–100]).
DiscussionThe S. pneumoniae detection is essential to supervise the epidemiological changes that occur in the incidence of IPD after the introduction of national immunization programs. The culture is the gold standard for confirmation of pneumococci. However this method has as major limitation that requires viable bacteria. For this reason culture has poor sensitivity in some samples as PF and after starting empirical antibiotic treatment. The main alternatives to microbiological culture are antigenic assays for detecting pneumococcal cell wall components (C-polysaccharides) common to all serotypes12 and molecular strategies (PCR). PCR is highly sensitive for S. pneumoniae detection13 and can be used after the antibiotic therapy. There are several targets for amplification of S. pneumoniae. The universal 16S PCR plus sequencing substantially improves the etiologic diagnosis of infectious pleural effusion11 and may be one good option for the detection of S. pneumoniae in clinical samples. This technique allows the simultaneously looking for a wide number of bacteria and facilitates the detection of infections with no previous knowledge of the etiologic agent. Nevertheless, it is prone to misidentify S. mitis as S. pneumoniae. In order to avoid these false positives results, more specific diagnosis strategies have been developed.14 Most PCR techniques designed for differential diagnosis of syndromes as pneumonia15 or meningitis16,17 uses a specific gene for S. pneumoniae together with a limited number of genes for different bacterial species in format of multiplex PCRs. Generally PCR assays directed to S. pneumoniae employ single genes associated to specific bacterial factors. The ply gene can be detected, besides S. pneumoniae, also in non pneumococcal streptococci, particularly S. pseudopneumoniae and S. mitis.1 The lytA gene may be as well found in other streptococci of the S. mitis group.18 And although lytA gene has been considered reference for pneumococcal PCR assays,1 the identification of pneumococci by lytA RT-PCR may lead to false results.19 It has been suggested that RT-PCR for combined detection of lytA and psaA would have high specificity in the diagnosis of pneumococcal infections.20 The plyA and lytA genes of S. pneumoniae form part of a pathogenicity island that is missing in ≈90% of S. mitis and completely absent in other streptococci of the mitis group (with the only exception of S. pseudopneumoniae isolates that consistently harbor it).18
Though the number of samples included is very limited, the results show that the clinical use of the multiplex RT-PCR technique to the lytA, plyA and psaA genes would be useful for the routine detection of S. pneumoniae in PF. The assay showed very good performance in the collection of the 81 PF samples, giving excellent sensitivity and specificity values. A weak point of this study lies on that the positivity S. pneumoniae PCR criteria were established according to previous clinical diagnosis results of lytA or 16S rRNA genes (no both at same time). In this sense, the multiplex lytA, plyA and psaA RT-PCR evaluates would really be more appropriate than the gold standards employed. The lowers sensitivity values obtained by the isolated detection of plyA and psaA genes may be due to that the main positivity criterion (the positivity to lytA by the in house assay) can be not necessary associated to the positivity to the other plyA or psaA genes. Although none of the three studied genes (lytA, plyA and psaA) constitutes an unequivocal marker of S. pneumoniae infection, in practical sense it is necessary to adopt an arbitrary consensus of positivity. The positivity of all the three genes is high specific and the positivity to only one high sensible. In fact, the use of three different genes can assure the specificity of the detection when some simultaneously positive results occur in the same sample and would solve troubles of misidentification.21 In this study 98.3% of the samples were positive at the same time for two genes (lytA and plyA) and 91.7% for three (lytA, plyA and psaA). No an assessment of the limit of detection of the in house lytA PCR assay neither of the commercial PCR evaluate technique were made in this study. However, according to the bibliography in witch are based both techniques,9 the limit of detection was equivalent to <10 genomic copies for lytA and plyA and 100 for psaA. In conclusion, although the number of studied samples is small, these results show that the use of the multiplex PCR for lytA, plyA and psaA genes may be a practical and useful option to identify S. pneumoniae directly in PF samples. The presentation as an easy kit and the short time of working (few hours) made suitable to clinical routine laboratories. Nonetheless, culture methods should continue to be used.22 The permanent possibility of S. pneumoniae emerging distinct serotypes, with variable antimicrobial susceptibility patterns, as are the candidates to new pneumococcal conjugate vaccine currently undergoing in clinical trials,23 made necessary an extensive strain based serotype surveillance.
Conflicts of interestThe authors declare that they have no conflicts of interest.
We thank Marisa Fernández, Mariluz Alvarez, Isabel Vadillo and Teresa Gómez for their excellent technical assistance.