In recent years, Streptococcus pneumoniae serotype 8 has become the most prevalent cause of invasive pneumococcal disease (IPD) in Madrid, Spain. The objective of this study was to characterize the invasive clones of S. pneumoniae serotype 8 in Madrid over the 2012–2015 period.
MethodsFrom January 2012 to December 2015, a total of 1543 invasive isolates were studied. Serotyping was carried out by Pneumotest-Latex agglutination and Quellung reaction. Susceptibilities to penicillin, erythromycin and levofloxacin were determined by the Etest®. All serotype 8 strains were typed by multilocus sequence typing (MLST) and by pulsed-field gel electrophoresis (PFGE).
ResultsTwo hundred and forty-eight (248) serotype 8 strains were detected (16.1%) and 243 of them were available for molecular typing. Nine sequence types (STs) by MLST (8-ST53, 8-ST63, 8-ST404, 8-ST1107, 8-ST989, 8-ST1110, 8-ST2231, 8-ST3544 and 8-ST4301), and nine PFGE profiles were identified (one corresponding to each ST). The 8-ST53 clone was the most widespread, and increased from 53.8% among all serotype 8 isolates in 2012, to 90.1% in 2015. In contrast, the 8-ST63 clone, resistant to levofloxacin and erythromycin, decreased from 30.8%, among all serotype 8 strains in 2012, to 5.0% in 2015.
ConclusionsThe increase in our region of S. pneumoniae serotype 8, not included in conjugated vaccines, occurred at the expense of the 8-ST53 clone. On the contrary, the 8-ST63 clone decreased. Since clone 8-ST63 has the theoretical advantage of its levofloxacin-erythromycin resistance in comparison to 8-ST53, the predominance of 8-ST53 over 8-ST63 is striking.
En los últimos años Streptococcus pneumoniae serotipo 8 ha sido la causa más prevalente de enfermedad neumocócica invasora (ENI) en la Comunidad de Madrid. El objetivo de este estudio fue caracterizar los clones invasores de S. pneumoniae serotipo 8 circulantes en Madrid ente los años 2012 y 2015.
MétodosSe estudiaron 1.543 cepas causantes de ENI aisladas entre enero de 2012 y diciembre de 2015. El serotipado se realizó mediante aglutinación con Pneumotest-Latex y reacción de Quellung. La determinación de la sensibilidad frente a penicilina, eritromicina y levofloxacino se realizó mediante Etest®. Las cepas del serotipo 8 se tipificaron por MLST (multi-locus sequence typing) y electroforesis en campo pulsado (PFGE).
ResultadosSe detectaron 248 cepas del serotipo 8 (16,1%) y 243 de ellas estuvieron disponibles para tipado molecular. Se identificaron 9 tipos de ST: 8-ST53, 8-ST63, 8-ST404, 8-ST1107, 8-ST989, 8-ST1110, 8-ST2231, 8-ST3544 y 8-ST4301, y 9 perfiles de PFGE (uno correspondiente a cada ST). El clon 8-ST53 fue el más prevalente dentro del serotipo 8 y aumentó del 53,8% en 2012 al 90,1% en 2015. Por el contrario el clon 8-ST63 asociado con resistencia a levofloxacino y eritromicina disminuyó del 30,8% en 2012 al 5,0% en 2015.
ConclusionesEl incremento del serotipo 8 en nuestra región, no cubierto por las actuales vacunas conjugadas, se produjo a expensas del clon 8-ST53. Inversamente, el clon 8-ST63 disminuyó. Dado que el clon 8-ST63 presenta sobre el 8-ST53 la ventaja teórica de su resistencia frente a levofloxacino y eritromicina, resulta llamativo el predominio de 8-ST53 sobre 8-ST63.
In November 2006, the 7-valent pneumococcal conjugate vaccine (PCV7) was included into the children's universal public funding immunization schedule of the Madrid Region (Spain).1 After its implementation, the invasive pneumococcal disease (IPD), affecting patients of all ages, was included in 2007 as a mandatory notifiable disease in the epidemiological surveillance system of the Region of Madrid. Since then, all invasive strains recovered in this Region, were sent to the Madrid Public Health Regional Laboratory for serotyping and antimicrobial susceptibility testing. In June 2010, the 13-valent pneumococcal conjugate vaccine (PCV13) replaced PCV7, but it was later excluded from the regional vaccination program in July 2012. After the universal funded vaccination withdraws, the PCV13 use continued in high-risk groups, while its administration in healthy infants only was available if purchased privately. Finally, PCV13 was reintroduced for the entire pediatric population in December 2014.1 Early after start to monitoring the IPD evolution in Madrid, during the period 2007–2009, serotype 8 was the first more frequent invasive non-PCV13 covered one (globally the sixth behind the PCV13 serotypes 1, 19A, 7F, 3 and 5) and was associated with pneumonia and bacteraemia, mainly in adults.2 The elevated rates of resistance to levofloxacin and erythromycin associated to serotype 8 during these years and its initial relationship to underlying risk factors as human immunodeficiency virus (HIV) infection was soon described.3,4 Over the last years, serotype 8 has become the most prevalent serotype causing IPD in Madrid in the general population.5 After the introduction of the PCV13, the incidence per 100,000 inhabitants decreased in Madrid from 10.52, in 2008–2010, to 7.09 in 2013–2015.5 However, the incidence of serotype 8 increased significantly from 0.60, in 2008–2010, to 1.07 in 2013–2015.5 The IPD incidence of other serotypes not included in conjugate vaccines (9N, 10A, 23B and 24F) also increased but at less magnitude.5 Although serotype investigation is paramount for understanding the pneumococcal epidemiology, molecular methods like multilocus sequence typing (MLST) are used in order to gain a deeper perspective of the circulation of pneumococci. Invasive serotype 8 isolates increased in Madrid from 2007 to 2009 by the spread of the levofloxacin, erythromycin and clindamycin resistant clone 8-ST63, with a S79F ParC mutation (in addition to other S81F, S81Y and E85K mutations in GyrA) and the constitutive macrolide-lincosamide-streptogramin B (cMLSB) phenotype.3,4 The objective of this study was the molecular characterization of invasive isolates of Streptococcus pneumoniae serotype 8 in Madrid (Spain), and to report the changes in circulating clones of this main serotype over the four-year period from 2012 to 2015.
Material and methodsFrom January 2012 to December 2015, all invasive pneumococcal isolates (one for every episode) from IPD cases of the Madrid Region (Spain) sent to the Madrid Public Health Regional Laboratory were studied. A total of 1543 invasive isolates from 1523 patients (754 cases younger than 18 years and 1284 adults) were recovered from 31 public and private hospitals. Identification of the capsular serotypes was carried out by the Pneumotest-Latex (Statens Serum Institut, Copenhagen, Denmark) and by Quellung reaction using commercial antisera (Statens Serum Institut, Copenhagen, Denmark). Susceptibilities to penicillin, erythromycin and levofloxacin were determined by the Etest® method (bioMerieux, France). MIC breakpoint interpretations were based on updated EUCAST-2018 standards.6 Pneumococcal isolates with penicillin MICs>0.06mg/l and >2mg/l were regarded as penicillin non-susceptible and penicillin resistant respectively. Strains with erythromycin MICs>0.5mg/l and levofloxacin MICs>2mg/l were considered resistant. MLST based on the aroE, gdh, gki, recP, spi, xpt, and ddl genes was performed, as described elsewhere,7 in a selected sample representative of S. pneumoniae serotype 8 strains. Sequence types (STs) were determined using the pneumococcal MLST database (https://pubmlst.org/spneumoniae/). Serotype 8 isolates were also analyzed by pulsed-field gel electrophoresis (PFGE). Genomic DNA was digested with SmaI enzyme8 and electrophoresis was done with ramped pulse times beginning with 1s and ending with 30s at 6V/cm for 22h. The temperature was set at 14°C using a CHEF-DR® III System (Bio-Rad Laboratories S.A., Madrid, Spain). The DNA banding patterns obtained by PFGE were analyzed with FPQuest Software (BioRad) and were interpreted according to the guidelines proposed by Tenover et al.9 The Dice coefficient of similarity was calculated, and the unweight pair group method with arithmetic averages (UPGMA) was used for cluster analysis. The percentage similarity between PFGE patterns was used to assess the relationship between the patterns. A cut-off at 90% similarity of the Dice coefficient was used to indicate identical PFGE types (this corresponds to approximately one band difference). PFGE patterns were related to representative international pneumococcal clones of the Pneumococcal Molecular Epidemiology Network (PMEN). The isolates were compared with six PMEN clones that were included in the PFGE analysis (Spain23F-1, Spain6B-2, Spain9V-3, Spain14-5, South Africa19A-13 and Taiwan23F-15). For the statistical analysis, categorical values were compared using chi-squared (χ2) and χ2 for linear trend or Fisher's test with it correspondent odds ratios (OR) if appropriate (p values <0.01 were considered significant). Incidences per 100,000 inhabitants were calculated according the census of the Madrid Region (6,498,560 inhabitants in 2012, 6,495,551 in 2013, 6,454,440 in 2014 and 6,436,996 in 2015).
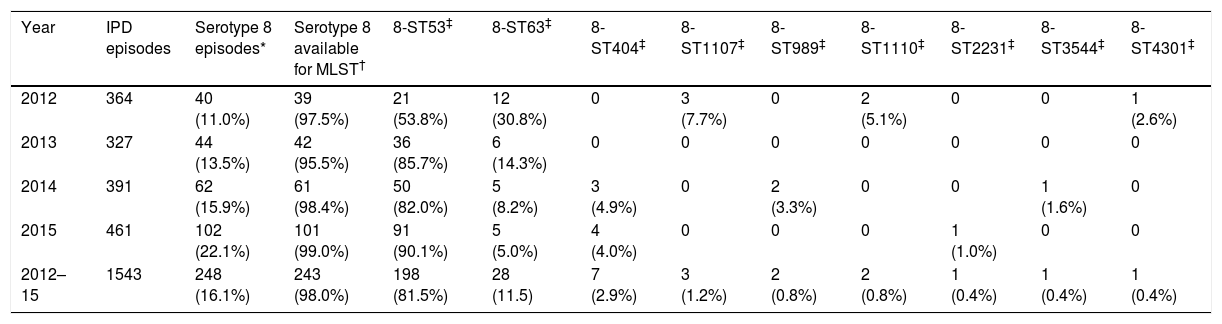
ResultsOverall, 18 serotypes comprised 77.3% out of the 1543 isolates as follows: serotype 8 (16.1%), 3 (8.9%), 19A (6.2%), 22F (4.7%), 11A (4.3%), 1 (4.2%), 9N (4.0%), 6C (3.3%), 10A (3.2%), 15A (3.1%), 23B (3.1%), 15B (2.5%), 24F (2.5%), 7F (2.5%), 31 (2.3%), 16F (2.3%), 12F (2.1%) and 35B (2.1%). The proportion of PCV13 non-covered serotypes increased significantly: 64.8% in 2012, 66.7% in 2013, 70.3% in 2014 and 79.2% in 2015 (p<0.001). The PCV13 non-covered serotype 8 was the most prevalent over the entire study period, and increased significantly (p<0.001) over the years 2012, 2013, 2014 and 2015 (40/364 [11.0%], 44/327 [13.5%], 62/391 [15.9%] and 102/461 [22.1%] respectively). Among the 248 serotype 8 detected strains, 243 were available for molecular typing. Nine different MLST serotype 8 patterns were detected: 8-ST53, 8-ST63, 8-ST404, 8-ST1107, 8-ST989, 8-ST1110, 8-ST2231, 8-ST3544 and 8-ST4301. Nine PFGE profiles were identified (each pulsotype corresponded to one MLST type). All ST 53 isolates, including those “resistant”, were clustered in the same PFGE type. Table 1 shows the MLST patterns of serotype 8 according to the year of detection. Serotype 8 isolates belonging to the 8-ST53 clone were the most widespread among the different clones of serotype 8, and increased significantly over the four years of the study 53.8% (2012), 85.7% (2013), 82.0% (2014) and 90.1% (2015) in comparison to other sequence types of serotype 8 (p<0.001). The 8-ST53 clone reached in 2015, at least, 19.7% (n=91; one serotype 8 isolate was not available for molecular typing) of the total of invasive pneumococci isolated in the Region of Madrid (n=461). By contrast, genotype 8-ST63 decreased significantly as follows: 30.8% (2012), 14.3% (2013), 8.2% (2014) and 5.0% (2015) (p<0.001). Hence, the incidence of genotype 8-ST53 increased (0.32 in 2012, 0.55 in 2013, 0.77 in 2014 and 1.41 in 2015). Inversely, the incidence of genotype 8-ST63 decreased (0.18 in 2012, 0.09 in 2013, 0.08 in 2014 and 0.08 in 2015). Only fifteen (6.2%) out of the 243 serotype 8 strains (fourteen 8-ST53 [4 isolated in 2013, 4 isolated in 2014 and 6isolated in 2015] and one 8-ST1107 [isolated in 2012]) were isolated from children younger than 18 years old.
Serotype 8 MLST patterns according to the year of detection.
| Year | IPD episodes | Serotype 8 episodes* | Serotype 8 available for MLST† | 8-ST53‡ | 8-ST63‡ | 8-ST404‡ | 8-ST1107‡ | 8-ST989‡ | 8-ST1110‡ | 8-ST2231‡ | 8-ST3544‡ | 8-ST4301‡ |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2012 | 364 | 40 (11.0%) | 39 (97.5%) | 21 (53.8%) | 12 (30.8%) | 0 | 3 (7.7%) | 0 | 2 (5.1%) | 0 | 0 | 1 (2.6%) |
| 2013 | 327 | 44 (13.5%) | 42 (95.5%) | 36 (85.7%) | 6 (14.3%) | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 2014 | 391 | 62 (15.9%) | 61 (98.4%) | 50 (82.0%) | 5 (8.2%) | 3 (4.9%) | 0 | 2 (3.3%) | 0 | 0 | 1 (1.6%) | 0 |
| 2015 | 461 | 102 (22.1%) | 101 (99.0%) | 91 (90.1%) | 5 (5.0%) | 4 (4.0%) | 0 | 0 | 0 | 1 (1.0%) | 0 | 0 |
| 2012–15 | 1543 | 248 (16.1%) | 243 (98.0%) | 198 (81.5%) | 28 (11.5) | 7 (2.9%) | 3 (1.2%) | 2 (0.8%) | 2 (0.8%) | 1 (0.4%) | 1 (0.4%) | 1 (0.4%) |
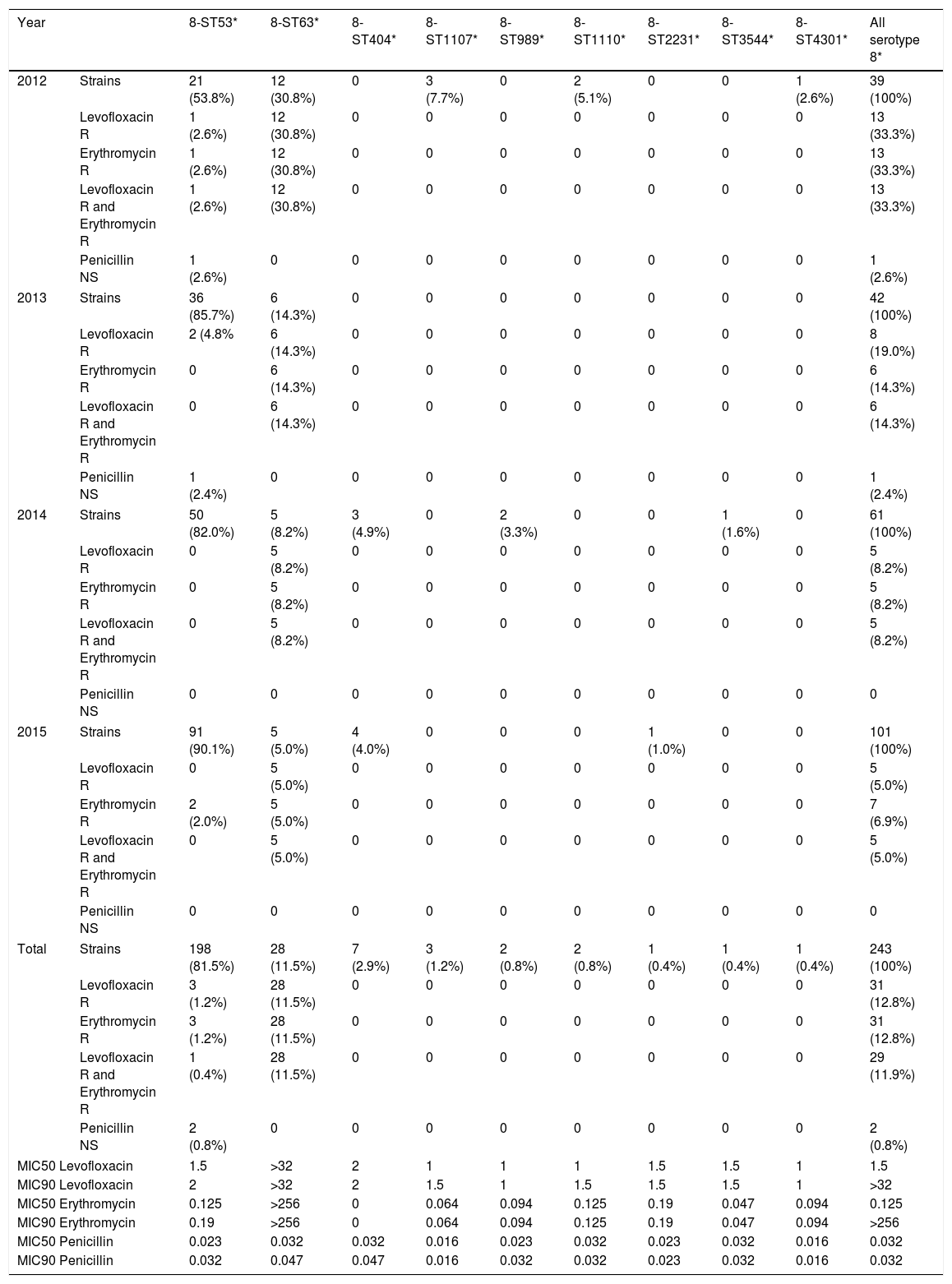
Concerning antimicrobial resistance, the rates of S. pneumoniae isolates non-susceptible to penicillin, resistant to levofloxacin, and resistant to erythromycin in Madrid over the period 2012–2015 remained stable: the percentages of penicillin non-susceptible and penicillin-resistant isolates 26.6% and 1.6% in 2012, 32.7% and 3.4% in 2013, 26.6% and 5.6% in 2014 and 26.5% and 3.9% in 2015; the figures for erythromycin resistance in 2012, 2013, 2014, and 2015 were 23.4%, 23.5%, 21.0%, and 21.3% respectively; and for levofloxacin resistance in 2012, 2013, 2014, and 2015 were 6.9%, 8.9%, 6.9% and 5.9% respectively. Sixty two (4.0%) strains were co-resistant to erythromycin and levofloxacin and belonged to serotypes 8 (48.4%), 15A (11.3%), 19A (9.7%), 9V (8.1%), 14 (6.5%), 24F (4.8%), 23A (3.2%), 6C (3.2%), 12F (1.6%), 15C (1.6%) and serogroup 33 (1.6%). The proportions of erythromycin and levofloxacin co-resistant strains by year among serotype 8 and other serotypes were 14/5 in 2012, 6/8 in 2013, 5/8 in 2014 and 5/11 in 2015 (respectively OR=1, OR=0.27, OR=0.22 and OR=0.16; χ2 for linear trend p<0.05). Considering the total of strains of all serotypes, the concomitant co-resistance to erythromycin and levofloxacin remained stable with no significant changes over the period analyzed (5.2% in 2012, 4.3% in 2013, 3.3% in 2014 and 3.5% in 2015).
Among serotype 8 isolates, the distribution of MLST patterns according to its antimicrobial susceptibility during the study period is shown in Table 2. Only genotypes 8-ST53 and 8-ST63 were detected in all years. Levofloxacin and erythromycin MIC90 among type 8-ST63 were >32 and >256mg/l respectively, and this clone was significantly associated to resistance and co-resistance to levofloxacin and erythromycin. None of the serotype 8 isolates were penicillin resistant. Two 8-ST53 isolates showed intermediate susceptibility to penicillin (MICs 0.094mg/L and 0.125mg/L). The remaining serotype 8 isolates (8-ST63. 8-ST404. 8-ST1107. 8-ST989. 8-ST1110. 8-ST2231. 8-ST3544 and 8-ST4301) were susceptible to penicillin.
Classification of serotype 8 MLST patterns according to its antimicrobial susceptibility along the study period.
| Year | 8-ST53* | 8-ST63* | 8-ST404* | 8-ST1107* | 8-ST989* | 8-ST1110* | 8-ST2231* | 8-ST3544* | 8-ST4301* | All serotype 8* | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 2012 | Strains | 21 (53.8%) | 12 (30.8%) | 0 | 3 (7.7%) | 0 | 2 (5.1%) | 0 | 0 | 1 (2.6%) | 39 (100%) |
| Levofloxacin R | 1 (2.6%) | 12 (30.8%) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 13 (33.3%) | |
| Erythromycin R | 1 (2.6%) | 12 (30.8%) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 13 (33.3%) | |
| Levofloxacin R and Erythromycin R | 1 (2.6%) | 12 (30.8%) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 13 (33.3%) | |
| Penicillin NS | 1 (2.6%) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 (2.6%) | |
| 2013 | Strains | 36 (85.7%) | 6 (14.3%) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 42 (100%) |
| Levofloxacin R | 2 (4.8% | 6 (14.3%) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 8 (19.0%) | |
| Erythromycin R | 0 | 6 (14.3%) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 6 (14.3%) | |
| Levofloxacin R and Erythromycin R | 0 | 6 (14.3%) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 6 (14.3%) | |
| Penicillin NS | 1 (2.4%) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 (2.4%) | |
| 2014 | Strains | 50 (82.0%) | 5 (8.2%) | 3 (4.9%) | 0 | 2 (3.3%) | 0 | 0 | 1 (1.6%) | 0 | 61 (100%) |
| Levofloxacin R | 0 | 5 (8.2%) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5 (8.2%) | |
| Erythromycin R | 0 | 5 (8.2%) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5 (8.2%) | |
| Levofloxacin R and Erythromycin R | 0 | 5 (8.2%) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5 (8.2%) | |
| Penicillin NS | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| 2015 | Strains | 91 (90.1%) | 5 (5.0%) | 4 (4.0%) | 0 | 0 | 0 | 1 (1.0%) | 0 | 0 | 101 (100%) |
| Levofloxacin R | 0 | 5 (5.0%) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5 (5.0%) | |
| Erythromycin R | 2 (2.0%) | 5 (5.0%) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 7 (6.9%) | |
| Levofloxacin R and Erythromycin R | 0 | 5 (5.0%) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5 (5.0%) | |
| Penicillin NS | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Total | Strains | 198 (81.5%) | 28 (11.5%) | 7 (2.9%) | 3 (1.2%) | 2 (0.8%) | 2 (0.8%) | 1 (0.4%) | 1 (0.4%) | 1 (0.4%) | 243 (100%) |
| Levofloxacin R | 3 (1.2%) | 28 (11.5%) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 31 (12.8%) | |
| Erythromycin R | 3 (1.2%) | 28 (11.5%) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 31 (12.8%) | |
| Levofloxacin R and Erythromycin R | 1 (0.4%) | 28 (11.5%) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 29 (11.9%) | |
| Penicillin NS | 2 (0.8%) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 (0.8%) | |
| MIC50 Levofloxacin | 1.5 | >32 | 2 | 1 | 1 | 1 | 1.5 | 1.5 | 1 | 1.5 | |
| MIC90 Levofloxacin | 2 | >32 | 2 | 1.5 | 1 | 1.5 | 1.5 | 1.5 | 1 | >32 | |
| MIC50 Erythromycin | 0.125 | >256 | 0 | 0.064 | 0.094 | 0.125 | 0.19 | 0.047 | 0.094 | 0.125 | |
| MIC90 Erythromycin | 0.19 | >256 | 0 | 0.064 | 0.094 | 0.125 | 0.19 | 0.047 | 0.094 | >256 | |
| MIC50 Penicillin | 0.023 | 0.032 | 0.032 | 0.016 | 0.023 | 0.032 | 0.023 | 0.032 | 0.016 | 0.032 | |
| MIC90 Penicillin | 0.032 | 0.047 | 0.047 | 0.016 | 0.032 | 0.032 | 0.023 | 0.032 | 0.016 | 0.032 | |
In the past, the prevalence of S. pneumoniae serotype 8 in Spain was low but constant over decades.10 Currently, in countries of Western Europe in which the PCV13 vaccine is used, serotype 8 is one of the most frequently recovered,11–13 and in England, in 2013 and 2014 it was the predominant invasive serotype detected.14 This serotype is especially important as a cause of bacteremic pneumonia in adults,15 while its specific contribution to pediatric IPD cases in the PCV13 era remains low.16 Serotype 8 is also increasing in countries that use the 10-valent pneumococcal conjugate vaccine (PCV10).17 During the period from 2012 to 2015, we observed a dramatic rise in the prevalence of serotype 8 in Madrid, from 11.0% in 2012 to 22.1% in 2015. This increase occurred mainly in the adult population.
The serotype is a key determinant of IPD potential and prevalence, however, analysis by PFGE and MLST typing, contribute to the genetic characterization and understanding of their spread. In this series, among serotype 8 isolates, we identified nine different STs corresponding to nine PFGE pulsotypes. The coincidence of PFGE profiles with STs demonstrates high molecular homogeneity of isolates belonging to the same ST. In this setting, the increase of serotype 8 in Madrid was associated to the clonal expansion and success of 8-ST53. In terms of incidence, the 8-ST53 clone increased from 0.32 per 100,000 inhabitants in 2013 to 1.41 in 2015, while 8-ST63 dropped from 0.18 per 100,000 inhabitants in 2012 to 0.08 in 2015. In a prospective multicenter study collecting adult IPD episodes, serotype 8 decreased after the period 2012–2013 in relation to a specific decline of the 8-ST63 clone.18 Pneumococcal serotype 8 has been associated with outbreaks,19,20 and the 8-ST53 clone has been linked to fatal pneumococcal disease in residents of a shelter for homeless men with history of excess alcohol consumption.21 Studies in Scotland performed 5 years before the introduction of PCV7 showed a raise in 8-ST53 cases.22 In addition, the 8-ST53 clone was the main non PCV13 recovered clone among bacteremic episodes of pneumonia in patients with chronic obstructive pulmonary disease during the first years of this century in Barcelona.23 In last years, this clone also increased significantly among adult cases in Portugal.24
The 8-ST63 clone emerging in Madrid before the PCV13 use, corresponded to a capsular switching event of the Sweden15A-ST63 clone which acted as a recipient of capsular DNA from the 8-ST53 strain which acted as a donor.25 The subsequent rise of the 8-ST53 clone, detected after 2012, seemed to be the result of the expansion of this (similar to this that play the role of donor in the ST63 switching). The clonal changes detected in serotype 8 were matched to variations in antimicrobial susceptibility profiles. The emergence and dissemination of multi-drug resistant pneumococcal clones are generated by the adaptation of the naturally residing human upper respiratory tract strains that transfer core genome and accessory resistance determinants to other highly transformable S. pneumoniae commensals, in response to the antibiotic external pressure.26 Moreover, vaccination programs may provide a selection pressure for virulent genotypes to switch capsules and escape the vaccine coverage. Since the 8-ST63 clone has, over the 8-ST53 clone, the theoretically ecological advantage of its levofloxacin–erythromycin resistance, the replace of 8-ST53 in place of 8-ST63 is striking. However, in this study we detected the 8-ST63 clone replacement by its precursor (and DNA donor) 8-ST53 clone. While both clones, 15A-ST63 recipient (resistant) and 8-ST53 donor (susceptible), are not included in conjugate vaccines, the decrease of levofloxacin–erythromycin resistance associated to the decrease of 8-ST63 switching was an independent event of the use of PCV13 vaccine. Although the proportion of levofloxacin and erythromycin co-resistant serotype 8 isolates decreased significantly over the study period, overall, this co-resistance decrease was compensated by the rise of other levofloxacin and erythromycin co-resistant serotypes (15A, 19A, 9V, 14, 24F, 23A, 6C, 12F, 15C and serogroup 33). It is difficult to know why the widely distributed levofloxacin and erythromycin resistant 8-ST63 clone was surpassed by the susceptible 8-ST53 clone. This fact is even more complex since the emerging 8-ST53 clone has been proposed as the genetic precursor of the withdrawing 8-ST63. It would be of interest to know the advantages of 8-ST53 against 8-ST63. Although under certain circumstances, antimicrobial resistance could help microorganisms to spread, in some cases, the acquisition of antibiotic resistance or virulence genes could reduce bacterial fitness making resistant bacteria unable to compete with the wild-type susceptible.
At present, serotype 8 has become the most frequent pneumococcal serotype isolated from IPD cases in the general population of our region. In this study none of the 28 episodes caused by the 8-ST63 clone occurred in children under 15 years. The increase of serotype 8 in Spain could compromise the ability of current PCVs to prevent IPD especially in adult patients. The 23-valent pneumococcal polysaccharide vaccine (PPV23) covers this capsular type. Since 2005, the PPV23 has been recommended in the Region of Madrid and financed by the public health system for all adults aged 60 years and over.27 The cumulative coverture of the PPV23 in patients older than 59 years in Madrid during 2015 was 62.1%.28 The vaccine effectiveness of PPV23 for IPD has been estimated in 64.2%.28 In Madrid in 2018, a single dose of PCV13 was recommended for adults ≥60 years without risk factors or comorbidities and also for adults ≥18 years old with chronic conditions. Some conjugate vaccine prototypes, including serotype 8, are in development. The new candidate 15-valent pneumococcal conjugate vaccine (PCV15) adds to PCV13 the serotypes 22F and 33F, but does not include serotype 8.29 The recently patented 20-valent pneumococcal conjugate vaccine (PCV20) incorporates, besides 22F and 33F, the serotypes 8, 10A, 11A, 12F and 15B.30 The inclusion of the serotype 8 could be crucial in our region for preventing IPD mainly in adults.
In conclusion, in this study, S. pneumoniae serotype 8, non-covered by conjugated vaccines, was the most prevalent in the general population and increased annually. This high prevalence was concentrated in adulthood. The increase of this serotype occurred at the expense of the widespread of the 8-ST53 clone that comprised almost 20% of the 2015 IPD cases in the Region of Madrid. In contrast, the levofloxacin–erythromycin co-resistant clone 8-ST63 decreased.
Conflict of interestJuan Carlos Sanz has received assistance from Pfizer for attending to scientific meetings. The others authors declare that they have no conflict of interest.