Brucellosis is a zoonotic infection transmitted to humans from infected animals by ingestion of food products (such as unpasteurized dairy products) or by contact with tissues or fluids. It is the most common zoonosis worldwide and is an important public health problem in many developing countries.1,2Brucella species are oxidase positive, catalase positive, Gram-negative coccobacilli causing brucellosis. Four Brucella species (B. abortus, B. melitensis, B. canis, B. suis) are known to cause disease in humans, however, most human cases are caused by B. melitensis. The matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI TOF- MS) provides fast, easy to perform and cost-effective diagnosis in clinical microbiology laboratories.3 Identification of Brucella spp. is not possible with MALDI-TOF MS, since this genus was not represented in the databases of the two main MALDI-TOF MS system manufacturers (such as bioMérieux and Bruker)4,5 and this may cause the misdiagnosis of brucellosis. In this letter, we present three cases in order to draw attention to the misidentification of Brucella melitensis as Ochrobactrum daejeonense by MALDI-TOF MS (Bruker, Germany) in our clinical microbiology laboratory.
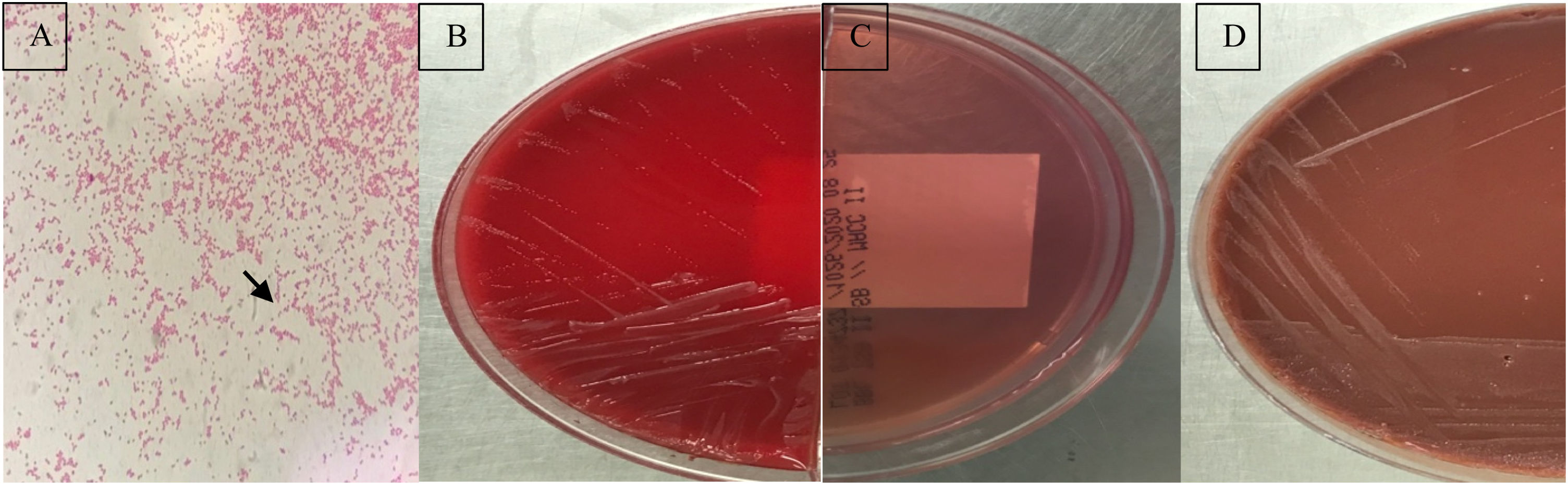
The first case was a 3-year-old female patient presenting with a complaint of intermittent fever and night sweats for a month. The patient had fever (38.5°C) as well as abdominal pain and weight loss. She had a history of consuming raw dairy products. The second case was a 44-year-old male presenting with weakness, chills and loss of appetite. The patient, who was engaged in animal husbandry, had a draining wound on his foot one month before. The third case was a 55-year-old male patient presenting fever, arthralgia and night sweats. He was diagnosed with brucellosis two years before, as stated in his medical history. Blood samples of these patients were sent to Hacettepe University clinical microbiology laboratory with the pre-diagnosis of brucellosis. Aerobic and anaerobic blood cultures were incubated in the Bactec FX (Becton-Dickinson, USA) automated blood culture system. Single, tiny gramnegative coccobacilli were observed in routine Gram staining made from the blood culture bottle giving a positive signal (Fig. 1A). The samples were inoculated on sheep blood, MacConkey, and chocolate agar. Growth on blood and chocolate agars showed non-hemolytic, transparent, flat, small colonies with the absence of growth on MacConkey agar (Fig. 1B–D). All three isolates tested positive for oxidase, catalase and urease, and were identified as Ochrobactrum daejeonense by MALDI-TOF MS (Bruker MALDI Biotyper, USA) with score values of 1.7, indicating genus-level identification but then, the colonies tested positive in the slide agglutination test using polyvalent Brucella spp. antiserum. Brucella spp. antibody titers were 1/1280, as determined with the Metser Coombs Brucella Test (Metserlab Biological Products, Turkey) in serum samples taken simultaneously from all three cases. The positivity of serological tests for brucellosis was an important clue for the possibility of a misidentification.
The definitive identification was further confirmed by polymerase chain reaction-based amplification of 16S rRNA and sequencing. DNA was extracted using the MasterPure DNA purification kit following the manufacturer's recommendations, with a modification of lysis as described by Wu et al.6 The amplification of the 1349-bp location of the 16S rRNA gene was performed using 27F 5′ AGAGTTTGATCMTGGCTCAG 3′ and 1492R 5′ TACGGYTACCTTGTTACGACTT primers. The cycles were: initial denaturation at 95°C for 5min, 40 cycles at 95°C for 20s, annealing at 57°C for 45s, extension 72°C for 1min, and final extension 72°C for 5min. The amplified products were run and viewed in a 1.5% agarose gel (Sigma, St. Louis, MO, USA). Sequencing was performed using BigDy Terminator V3.1 Cycle Sequencing Kit (Applied Biosystems, MA, USA). DNA sequences of the purified products were identified using ABI Prism 3700 Genetic Analyzer (AppliedBiosystems). The isolates were identified comparing the DNA reference isolates with data stored in the GenBank using the Basic Local Alignment Search Tool (BLAST version 2.0; http://www.ncbi.nlm.nih.gov/BLAST) program. A phylogenetic tree analysis was created using ClustalW MegAlign [https://www.ncbi.nlm.nih.gov/genbank/]. According to DNA sequence analysis 16S rRNA was 99% compatible with Brucella melitensis.
The family Brucellaceae consists of seven genera (Brucella, Ochrobactrum, Crabtreella, Daeguia, Mycoplana, Paenochrobactrum, and Pseudochrobactrum). Brucella and Ochrobactrum are closely related genera of the Brucellaceae family. Brucella spp. have been formerly misidentified as Ochrobactrum antropi with several automated systems.7–10 MALDI-TOF MS is a reliable and rapid method for bacterial identification. Some databases used for this purpose lack reference profiles for Brucella species, due to its potential bioterrorism application. Bruker has a cooperation with the Centers for Disease Control (CDC) since 2016 (https://microbenet.cdc.gov/) so, people attempting the identification of bioterrorism microorganisms can be tracked. It would be useful for laboratory workers to have also this information as an alternative to the identification obtained directly from the mass spectrometer. When MALDI-TOF users obtain the identification “Ochrobactrum spp.” and suspect misidentifications of Brucella spp., this suspicion can be confirmed using the CDC database, since Brucella species are still an important pathogen in wide areas around the world.
In this letter, three cases of B. melitensis were misidentified as Ochrobactrum daejeonense by MALDI-TOF MS in our clinical microbiology laboratory. The identification of the three isolates described in this letter was confirmed by PCR and sequencing of the 16S rRNA. There are several reasons why Brucella species are often misidentified in clinical laboratories. First, Brucella spp. infections are relatively rare, which leads to a lack of experience with this organism in clinical laboratories in some countries. Second, different bacterial automated identification systems are unable to correctly identify Brucella species, which misleads technical staff. Nevertheless, clinical microbiology laboratories in countries where brucellosis is endemic must be careful while using automated bacterial identification systems for identification of Brucella spp. as in our country.
In summary, accurate and rapid identification of Brucella species is most important for the early initiation of appropriate treatment. If there is a clinical suspicion of brucellosis, we suggest that the Ochrobactrum daejeonense or any other Ochrobactrum species results obtained with MALDI-TOF MS should be confirmed by additional tests such as biochemical, serological or molecular methods. These results should be evaluated with the correspondent clinical demonstration. Countries where brucellosis is endemic must be aware of the limitations of the MALDI-TOF MS for Brucella spp. identification. On the other hand, misidentification of Brucella spp. in the laboratory carries a high risk of laboratory-acquired infection due to aerosol generation and exposure among the laboratory personnel.11Brucella spp. should be handled under biosafety level 3 (BSL-3) conditions. Misidentification of Brucella spp. isolates as Ochrobactrum spp. can cause incorrect management of Brucella spp. cultures outside BSL3 laboratories.
Ethical approvalNot required.
FundingNo funding sources.
Competing interestsNone declared.