Herpes simplex virus (HSV) is a wide spread pathogen with symptoms during infection ranging from completely asymptomatic to a combination of mild symptoms.1 In critically ill patients HSV can cause life-threatening pulmonary disease.2 Cell culture is the gold standard for the diagnosis of HSV, but 2–3 days are needed to detect the cytopathic effect.1 New technologies, especially polymerase chain reaction (PCR) allows more rapid, albeit more costly, detection of the virus.3 Loop-mediated isothermal amplification (LAMP)4 has shown to be comparable with culture and PCR for the detection of HSV in genital lesions.5 The aim of this study was to evaluate the efficacy of LAMP compared to conventional culture to detect HSV-1 in bronchoalveolar lavage fluid (BALF).
A total of 50 BALF samples were included from patients with clinically suspected respiratory infection. The samples were routinely cultured in human fibroblast cells and monitored for up to 3 weeks to determine the presence of HSV by cytopathic effect observation and immunofluorescent detection of the antigen using monoclonal antibodies (MicroTrak® HSV1/HSV2 Direct Specimen Identification/Typing Test Kit, TrinityBiotech, Ireland).
A set of six previously described primers against the gene encoding the HSV-1 glycoprotein G (gG) were used,6 comprising two outer, two inner and two loop primers. The LAMP reaction was performed with 25μL of the reaction mixture: 2.5μL mix of primers (final concentration 0.2μM for outer primers, 1.6μM for inner primers and 0.4μM for loop primers), 15μL Isothermal Master Mix (Optigene, United Kingdom), 2.5μL PCR grade water and 5.0μL DNA/sample. The reaction was conducted in a Versant kPCR System (Siemens) at 65°C for 40minutes and the LAMP product detected by real time fluorescence. For each sample three reactions were performed: directly from the sample, after boiling 50μL of sample for 10minutes and after automatically extracting the DNA using the DSP Virus/Pathogen Midi kit (Qiagen, Germany) in a QIAsymphony automated platform (Qiagen, Germany). The specificity of the LAMP assay was evaluated using extracted DNA from 3 positive samples of HSV-2, varicella zoster virus (VZV), Epstein-Bar virus (EBV), cytomegalovirus (CMV) and herpesvirus 6 (HV6). Molecular detection of HSV-1 was performed by real-time quantitative PCR (rtPCR) (HSV1 Q-PCR Alert AmpliMIX, ELITechGroup NANOGEN®, Italy) with extracted DNA in case of discrepancies between culture and LAMP.
Culture detected 27 positive and 23 samples were negative, whereas LAMP detected HSV-1 in 23 samples in which DNA was extracted and in 21 samples in which LAMP was performed after boiling or directly with the sample. LAMP products were not detected with the DNAs of HSV-2, VZV, EBV, CMV and HV6. After DNA extraction, LAMP only failed to detect HSV-1 in five cases in which rtPCR was also negative (two cases) or presented late threshold cycles (three cases). In one case with negative culture, all variants of LAMP and rtPCR were positive.
Excluding the two samples with negative results for rtPCR and LAMP, the concordance observed between culture and LAMP was 92.0% (44/48, kappa coefficient of 0.834) for LAMP after extracting the DNA and 88.0% (42/48), kappa coefficient of 0.751) for LAMP after boiling or directly with the sample. The sensitivity of LAMP was 88.0% (22/25) after DNA extraction and 80.0% (20/25) when performed after boiling or using directly the sample (Table 1).
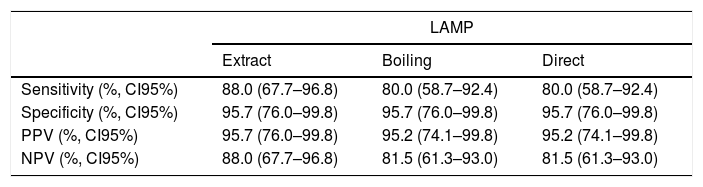
Sensitivity, specificity, positive predictive value and negative predictive value of LAMP (using DNA obtained by extraction or performed after boiling or directly with the sample) for HSV-1 detection from bronchoalveolar samples.
| LAMP | |||
|---|---|---|---|
| Extract | Boiling | Direct | |
| Sensitivity (%, CI95%) | 88.0 (67.7–96.8) | 80.0 (58.7–92.4) | 80.0 (58.7–92.4) |
| Specificity (%, CI95%) | 95.7 (76.0–99.8) | 95.7 (76.0–99.8) | 95.7 (76.0–99.8) |
| PPV (%, CI95%) | 95.7 (76.0–99.8) | 95.2 (74.1–99.8) | 95.2 (74.1–99.8) |
| NPV (%, CI95%) | 88.0 (67.7–96.8) | 81.5 (61.3–93.0) | 81.5 (61.3–93.0) |
LAMP: loop-mediated isothermal amplification; PPV, positive predictive value; NPV, negative predictive value; CI95%: 95% confidence interval.
The average time to achieve a positive result was 75.7minutes after DNA extraction (60min for extraction plus 15.7min for LAMP), 30.6min for LAMP after boiling (10min for boiling plus 20.6min for LAMP), and 22.8min for LAMP performed directly with the sample versus three to five days for virus culture.
LAMP assays are emerging as a good and inexpensive alternative for the diagnosis of HSV-1 infections. Previous studies have evaluated LAMP to detect HSV-1 in vitreous,7 cutaneous5,8 and cerebrospinal fluid samples,9 but this is the first study to evaluate the use of a LAMP in respiratory samples. Our results showed very good concordance between culture and LAMP. Performing LAMP directly from the sample saved time and showed similar results compared to LAMP performed in extracted DNA. Discordances with positive culture and negative rtPCR might be explained by the freeze and thaw cycles (DNA was probably degraded). On the other hand, the case with negative culture and positive LAMP and rtPCR could be explained by the virus not being viable. This is not the first time that the results of LAMP performed with and without DNA extraction have shown similar results.8 By avoiding the extraction step, LAMP can be performed in short time and is less expensive. The presence of HSV-1 in BALF is related to patient outcomes,10 including increased mortality. However, few laboratories include the detection of HSV-1 in respiratory samples from critically ill patients thereby leading to possible misdiagnosis of infection. One of the limitations of this study is the lack of analytical sensitivity, but the main objective is to detect those patients with higher viral loads and worse potential outcome. The results of this study show that the LAMP assay can be used as a rapid screening tool to detect HSV-1 in BALF from these patients.
FundingsThis work was supported by Ajut a la Recerca “Clínic-La Pedrera” 2016 (PEP:HB-16-JF-VG-C) and grant 2014SGR0653 from the Departament de Universitats, Recerca i Societat de la Informació de la Generalitat de Catalunya, by the Ministerio de Economía y Competitividad, Instituto de Salud Carlos III, co-financed by European Regional Development Fund (ERDF) “A Way to Achieve Europe,” the Spanish Network for Research in Infectious Diseases (REIPI RD16/0016/0010).