The ability of Spanish microbiology laboratories to (a) determine antimicrobial susceptibility (AS), and (b) correctly detect the vancomycin resistance (VR) phenotype in vancomycin-resistant Enterococcus spp. (VRE) was evaluated.
MethodsThree VRE isolates representing the VanA (E. faecium), VanB (E. faecium) and VanC (E. gallinarum) VR phenotypes were sent to 52 laboratories, which were asked for: (a) AS method used; (b) MICs of ampicillin, imipenem, vancomycin, teicoplanin, linezolid, daptomycin, ciprofloxacin, levofloxacin and quinupristin–dalfopristin, and high-level resistance to gentamicin and streptomycin; (c) VR phenotype.
Results(a) The most frequently used system was MicroScan; (b) according to the system, the highest percentage of discrepant MICs was found with gradient strips (21.3%). By antimicrobial, the highest rates of discrepant MICs ranged 16.7% (imipenem) to 0.7% (linezolid). No discrepant MICs were obtained with daptomycin or levofloxacin. Mayor errors (MEs) occurred with linezolid (1.1%/EUCAST) and ciprofloxacin (5.0%/CLSI), and very major errors (VMEs) with vancomycin (27.1%/EUCAST and 33.3%/CLSI) and teicoplanin (5.7%/EUCAST and 2.3%/CLSI). For linezolid, ciprofloxacin, and vancomycin, discrepant MICs were responsible for these errors, while for teicoplanin, errors were due to a misassignment of the clinical category. An unacceptable high percentage of VMEs was obtained using gradient strips (14.8%), especially with vancomycin, teicoplanin and daptomycin; (c) 86.4% of the centers identified VanA and VanB phenotypes correctly, and 95.0% the VanC phenotype.
ConclusionMost Spanish microbiology laboratories can reliably determine AS in VRE, but there is a significant percentage of inadequate interpretations (warning of false susceptibility) for teicoplanin in isolates with the VanB phenotype.
Se evaluó la capacidad de los laboratorios de microbiología españoles para: (a) determinar la sensibilidad antimicrobiana (SA); y (b) detectar correctamente el fenotipo de resistencia a vancomicina (FRV) en Enterococcus spp. resistente a vancomicina (ERV).
MétodosSe enviaron 3 aislados de ERV (E. faecium/VanA, E. faecium/VanB y E. gallinarum/VanC) a 52 laboratorios, a los que se les solicitó: (a) método de SA; (b) CMI de ampicilina, imipenem, vancomicina, teicoplanina, linezolid, daptomicina, ciprofloxacino, levofloxacino y quinupristina-dalfopristina y resistencia de alto nivel a gentamicina y estreptomicina; y (c) fenotipo de resistencia a vancomicina.
Resultados(a) El sistema más utilizado fue MicroScan; y (b) el mayor porcentaje de CMI discrepantes se produjo con las tiras de gradiente (21,3%). Las tasas más elevadas de CMI discrepantes variaron entre el 16,7% (imipenem) y el 0,7% (linezolid). Se produjeron errores mayores con linezolid (1,1%/EUCAST) y ciprofloxacino (5,0%/CLSI) y errores máximos con vancomicina (27,1%/EUCAST y 33,3% CLSI) y teicoplanina (5,7%/EUCAST y 2,3%/CLSI). Para linezolid, ciprofloxacino y vancomicina las CMI discrepantes fueron las responsables de estos errores, mientras que para teicoplanina los errores se debieron a una asignación errónea de la categoría clínica. Se obtuvo un alto porcentaje de errores máximos utilizando tiras de gradiente (14,8%), especialmente con vancomicina, teicoplanina y daptomicina; y (c) el 86,4% de los centros identificaron correctamente los fenotipos VanA y VanB y el 95,0% el fenotipo VanC.
ConclusiónLa mayoría de los laboratorios de microbiología españoles determinan de forma fiable la SA en ERV, pero existe un porcentaje significativo de interpretaciones inadecuadas (falsa sensibilidad) para teicoplanina en aislados con fenotipo VanB.
Enterococci are common opportunistic human pathogens able to cause a wide spectrum of infections, ranging from moderate (e.g. UTI) to severe (e.g. endocarditis), which are associated with high mortality rates, particularly in hospitalized immunocompromised patients with severe underlying diseases.1
The most clinically relevant enterococcal species are Enterococcus faecalis and E. faecium, which display intrinsic resistance to many antimicrobials, such as cephalosporins and aminoglycosides; E. faecium, included in the ESKAPE group of pathogens, is more frequently resistant to vancomycin and ampicillin than E. faecalis.1 Vancomycin is frequently used as first-line treatment for severe infections caused by ampicillin-resistant enterococci; however, vancomycin resistance (VR) is increasingly being reported worldwide. In Europe, according to the 2019 European Antimicrobial Resistance Surveillance Network (EARS-Net), VR among E. faecalis isolates remained low in most EU/EEA countries, whereas among E. faecium, the population-weighted mean percentage increased from 10.5% in 2015 to 17.3% in 2019.2 The main mechanism of VR in enterococci involves a substitution of the d-Alanine-d-Alanine in the peptidoglycan leading to variable expressions of VR. Most operons related to VR in enterococci (vanA, -B, -C, -D, -E, -G, -L, -M, and N) differ in their degree of VR, location (chromosome and plasmid), transferability, and phenotypic expression (constitutive or inducible). The vanA and vanB gene clusters are the most frequently implicated in VRE outbreaks. The vanA gene cluster is associated frequently with transposons (Tn), such as Tn1546, and it is formed by seven genes. The vanR (response regulator) and vanS (sensor kinase) gene products constitute the two-component system regulatory apparatus. The vanHXA gene cluster is essential for VR. The vanH and vanA code for a dehydrogenase that converts pyruvate to lactate (VanH), and for a ligase that forms d-Alanine-d-lactate dipeptide (VanA), which are implicated in the modification of peptidoglycan precursors, whereas the vanX (dipeptidase that cleaves d-Alanine-d-Alanine) and vanY (d,d-carboxypeptidase) are implicated in the hydrolysis and interruption of the pentapeptides. VanA-resistant strains show high-level resistance to vancomycin (MICs≥64mg/l) and teicoplanin (MICs, ≥16mg/l).3–5
The typical vanB operon has a similar genetic backbone to vanA. The vanHBX gene cluster is essential for VR and it can be associated to transposons such as Tn1547, Tn1549, and Tn5382. The conjugative Tn1549 is widely prevalent and it is mainly a chromosomal transposon. Isolates with the VanB resistance phenotype (vanB-1 and vanB-2 subtypes) are resistant to variable levels of vancomycin (MICs, 4–>1000mg/l) but are generally susceptible to teicoplanin. VanB strains however may also exhibit resistance to teicoplanin and thus be phenotypically indistinguishable from VanA strains.3–5
The vanC operon is genetically different from vanA and vanB, and it is typically present in “less virulent” enterococci than those carrying inducible vanA and vanB gene clusters. The vanC-resistant subtypes, vanC-1, vanC-2, and vanC-3, are known to be intrinsically present in E. gallinarum, E. casseliflavus, and E. flavescens, respectively, which display low-level resistance to vancomycin (MICs, 4–32mg/ml) and susceptibility to teicoplanin.3–5
Reliable antimicrobial susceptibility results and the recognition of VR phenotypes in the laboratory can directly impact outcome in VRE-infected patients because the treatment options available are limited and the probability of therapeutic failure is high.6,7
Unreliable antimicrobial susceptibility results have been related to several factors, such as species identification, type of acquired resistance mechanism, and the methodology used (broth microdilution versus disk diffusion).8–11 The clinical breakpoints applied, as well as the inducibility of resistance and the possibility of resistance development during treatment, can also affect the interpretation of susceptibility results.12–14 These challenges led us to perform the present study with the aim of evaluating the ability of Spanish clinical microbiology laboratories (a) to determine the antimicrobial susceptibility of VRE, and (b) to correctly detect different VR phenotypes.
Materials and methodsBacterial isolates, species identification and antimicrobial susceptibility testingThree Enterococcus spp. isolates (CC-01, CC-02 and CC-03) representing the VanA, VanB and VanC-1 VR phenotypes were selected for this study (Table 1). These VR phenotypes were characterized phenotypically, determining the MICs of vancomycin and teicoplanin (see below), and genetically, detecting the vancomycin resistance genes vanA, vanB and vanC-1 by PCR amplification and DNA sequencing with specific primers for vanA, vanB and vanC.12 All isolates were collected from patients admitted to the University Hospital Gregorio Marañón (Madrid, Spain).
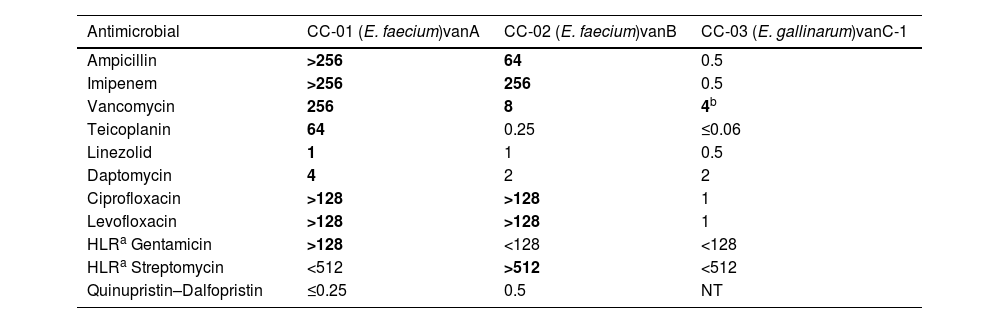
MICs (mg/l) of 11 antimicrobials against Enterococcus spp. isolates with different vancomycin resistance phenotypes.
| Antimicrobial | CC-01 (E. faecium)vanA | CC-02 (E. faecium)vanB | CC-03 (E. gallinarum)vanC-1 |
|---|---|---|---|
| Ampicillin | >256 | 64 | 0.5 |
| Imipenem | >256 | 256 | 0.5 |
| Vancomycin | 256 | 8 | 4b |
| Teicoplanin | 64 | 0.25 | ≤0.06 |
| Linezolid | 1 | 1 | 0.5 |
| Daptomycin | 4 | 2 | 2 |
| Ciprofloxacin | >128 | >128 | 1 |
| Levofloxacin | >128 | >128 | 1 |
| HLRa Gentamicin | >128 | <128 | <128 |
| HLRa Streptomycin | <512 | >512 | <512 |
| Quinupristin–Dalfopristin | ≤0.25 | 0.5 | NT |
NT: not tested.
MICs shown in bold correspond to the resistant clinical category according to EUCAST breakpoints, version 8.1, whereas MICs in normal type correspond to the susceptible category.
E. faecalis ATCC 29212 was used as quality control strain for antimicrobial susceptibility testing.
Bacterial identification, antimicrobial susceptibility and confirmation of the phenotype and genotype of VR were verified independently by two clinical microbiology reference laboratories: the Hospital Universitario Virgen Macarena (Seville, Spain), and the Hospital General Universitario Gregorio Marañón (Madrid, Spain). The isolates were identified using conventional microbiological tests and MALDI-TOF (Bruker Daltonics, Madrid, España). The antimicrobials tested were: ampicillin, imipenem, vancomycin, teicoplanin, linezolid, daptomycin, ciprofloxacin, levofloxacin, gentamicin (high-level resistance), streptomycin (high-level resistance) and quinupristin–dalfopristin. All antimicrobials were tested in duplicate at each reference center by broth microdilution and disk diffusion, according to EUCAST and CLSI guidelines.15,16 The 2018 EUCAST and CLSI breakpoints were used for the interpretation of clinical categories.15,16 The EUCAST breakpoints used for ciprofloxacin and levofloxacin were those reported for uncomplicated urinary tract infections. Quinupristin–Dalfopristin susceptibility results for isolate CC-03 (E. gallinarum vanC-1) were not included in the analysis because neither EUCAST nor CLSI have breakpoints for quinupristin–dalfopristin in enterococcal species other than E. faecium.
Study designIsolates were sent in Amies transport medium to 52 participating hospitals in October 2018. The instructions specified that isolates should be treated as blood culture isolates. Participating laboratories were requested to fill in an electronic form for each isolate, which included: (a) method used for AST; (b) MIC values of ampicillin, imipenem, vancomycin, teicoplanin, linezolid, daptomycin, ciprofloxacin, levofloxacin quinupristin–dalfopristin, and high-level resistance to gentamicin and streptomycin; (c) breakpoint used for clinical category interpretation (CLSI or EUCAST); and (d) inference or detection of the phenotype of resistance to vancomycin (VanA, Van B or VanC).
Data analysisThe analysis of results consisted of: (a) a descriptive analysis of AST methods, breakpoints applied, clinical category assigned; (b) an analysis of discrepancies in MIC values; (c) an analysis of category error rates [minor errors (mEs), major errors (MEs) and very major errors (VMEs); and (iv) the ability of participating laboratories to accurately infer or detect the phenotype of VR.17
It was considered that there was a discrepancy in the MIC value of any antimicrobial tested when the MIC provided by the participating laboratory was not within a single 2-fold dilution (±1 doubling dilution) of the reference MIC result.
ResultsFifty-one out of the 52 centers invited to participate in this study responded to the questions related to the type of AST system used, the MICs obtained for the antimicrobials tested, the clinical breakpoints applied, and the type of VR phenotype identified.
Type of AST systemOne thousand and nine MIC determinations were analyzed. The percentage distribution of MIC determinations obtained with each AST system used was: 60.2% (607/1009) with MicroScan WalkAway (Beckman Coulter Inc., Brea, CA, USA); 29.5% (298/1009) with Vitek 2 (bioMérieux, Marcy-l’Étoile, France); 5.9% (60/1009) with gradient strips (Etest®, bioMérieux); 2.7% (27/1009) using an in-house broth microdilution test; and 1.7% (17/1009) with Phoenix (BD Biosciences, Sparks, MD, USA).
Discrepancies in the MICsAnalysis of discrepant MICs according to AST system revealed that the gradient strip from bioMérieux was the least accurate method for AST, with 21.3% of discrepant MICs (Table 2).
Distribution of discrepancies in MIC values and category error rates for Enterococcus spp. using different antimicrobial susceptibility testing systems.
| AST system (no. of centers using the system/total no. of centers; %) | % Discrepant MICsa | Clinical category errors (%)b | |||||
|---|---|---|---|---|---|---|---|
| Overall | ME | VME | |||||
| EUCAST | CLSI | EUCAST | CLSI | EUCAST | CLSI | ||
| Gradient strips (19/51; 37.3%) | 21.3 | 14.8 | 12.1 | 0.0 | 0.0 | 14.8 | 12.1 |
| MicroScan (32/51; 62.7%) | 4.1 | 5.6 | 4.6 | 0.0 | 0.0 | 5.6 | 4.1 |
| Broth microdilution (2/51; 3.9%) | 3.7 | 3.5 | 3.0 | 0.0 | 0.5 | 3.5 | 3.0 |
| Vitek (16/51; 31.4%) | 1.7 | 0.0 | 4.8 | 0.0 | 0.0 | 0.0 | 4.8 |
| Phoenix (1/51; 2.0%) | 0.0 | 5.9 | 0.0 | 0.0 | 0.0 | 5.9 | 0.0 |
By antimicrobial, the percentages of discrepant MICs were 16.7% for imipenem, 13.0% for ampicillin, 9.2% for teicoplanin, 2.0% for vancomycin, 1.9% for quinupristin–dalfopristin, 1.4% for ciprofloxacin, 0.7% for linezolid, and no discrepancies in the MICs of daptomycin, levofloxacin, and HLR to gentamicin or streptomycin were observed (Table 3).
Distribution of discrepancies in MIC values and category error rates for Enterococcus spp. by antimicrobial.
| Antimicrobiala (no.) | % of discrepant MICsb | No. of MICs interpreted using breakpoints ofc | Category errorsc | ||||
|---|---|---|---|---|---|---|---|
| EUCAST | CLSI | MEs | VMEs | ||||
| EUCAST | CLSI | EUCAST | CLSI | ||||
| Imipenem (30) | 16.7 | 25 | 5 | 0.0 | 0.0 | 0.0 | 0.0 |
| Ampicillin (138) | 13.0 | 95 | 43 | 0.0 | 0.0 | 0.0 | 0.0 |
| Teicoplanin (152) | 9.2 | 107 | 45 | 0.0 | 0.0 | 27.1 | 33.3 |
| Vancomycin (150) | 2.0 | 106 | 44 | 0.0 | 0.0 | 5.7 | 2.3 |
| Quinupristin–dalfopristin (52) | 1.9 | 35 | 17 | 0.0 | 0.0 | 0.0 | 0.0 |
| Ciprofloxacin (73) | 1.4 | 53 | 20 | 0.0 | 5.0 | 0.0 | 0.0 |
| Linezolid (135) | 0.7 | 89 | 46 | 1.1 | 0.0 | 0.0 | 0.0 |
| Daptomycin (72) | 0.0 | 30 | 42 | 0.0 | 0.0 | 0.0 | 0.0 |
| Levofloxacin (91) | 0.0 | 60 | 31 | 0.0 | 0.0 | 0.0 | 0.0 |
| HLR to gentamicin (69) | 0.0 | 39 | 30 | 0.0 | 0.0 | 0.0 | 0.0 |
| HLR to streptomycin (47) | 0.0 | 20 | 27 | 0.0 | 0.0 | 0.0 | 0.0 |
The number of MIC determinations reported for each antimicrobial is in parentheses. All the antimicrobials were not tested using the five AST methods. HLR: high-level resistance.
With respect to clinical breakpoint, 65.3% of MICs reported were interpreted using EUCAST-2018 breakpoints, and 34.7% with CLSI-2018 breakpoints.
Regarding clinical category errors according to AST system (see Table 2), VMEs were observed with all the AST methods used, particularly gradient strips (14.8% with EUCAST breakpoints and 12.1% with CLSI). The percentage of MEs was 0.5% with CLSI, and no mEs were obtained.
With respect to the breakpoint used, the overall percentage of VMEs obtained was 5.3% with EUCAST and 4.6% with CLSI breakpoints. By contrast, the percentage of MEs were 0.2% with EUCAST and 0.3% with CLSI breakpoints, and no mEs were obtained with either EUCAST or CLSI. According to antimicrobial, clinical category errors were detected for teicoplanin, vancomycin, ciprofloxacin, and linezolid (Table 3). VMEs occurred only with teicoplanin (27.1% using EUCAST, 33.3% with CLSI) and vancomycin (5.7% with EUCAST, 2.3% with CLSI) whereas the MEs were observed with ciprofloxacin (5.0% using CLSI) and linezolid (1.1% using EUCAST) (Table 3).
For teicoplanin, the percentage of VMEs related to clinical categorization obtained was associated with use of AST system (62.1% MicroScan, 24.1% Vitek 2, 10.3% in-house broth microdilution, and 3.4% Phoenix), however these AST systems did not produce discrepant MIC values (all ≤4mg/l). On the contrary, the VMEs were related to erroneous interpretation of MIC results (see below). For vancomycin, and in contrast to teicoplanin, the percentage of VMEs was caused by discrepant MICs ≤4mg/l (MIC underestimation), obtained using MicroScan (83.3%) and gradient strips (16.7%).
The percentage of ciprofloxacin MEs was due to discrepant MICs of >2mg/l (MIC overestimation), obtained using MicroScan and interpreted following CLSI breakpoints (5.0%). Similarly, for linezolid, the percentage of MEs was caused by MIC discrepancies of >4mg/l (MIC overestimation) using MicroScan and EUCAST breakpoints (1.1%).
Identification of VR phenotypesThe VanA and VanB phenotypes were correctly identified by 86.4% of the centers. The VanC phenotype was reported by 95% of the centers, which reported this result as follows: intrinsic resistance to glycopeptides (10%), vanC phenotype (47%), vanC-1 phenotype (8%), and intrinsic resistance and VanC phenotype (33%). All clinical category errors obtained for vancomycin and teicoplanin (VMEs) occurred with the VanB isolate; for linezolid, the errors (MEs) occurred with the VanA isolate, and for ciprofloxacin (MEs), with the VanC isolate.
DiscussionAccurate AST in enterococci is essential for proper guidance of therapy, particularly in immunocompromised patients with severe infections, because of the high associated morbidity and mortality, and for surveillance of antimicrobial resistance.6,18,19
The use of unreliable MIC-based AST systems can lead to inaccurately or wrongly estimated MIC values, and to false-susceptible or false-resistant results, which could in turn lead to failure in detecting nosocomial outbreaks caused by VRE isolates, including those produced by hospital-associated linages and high risk clones, such as clade A of E. faecium and clusters PP2, PP6, PP7, PP8 and PP20 of E. faecalis.20,21
The results of the present study show that discrepant MICs and category errors, whether or not associated with erroneous MIC results, are related to the AST system used, the antimicrobial agent, and the VR phenotype.
The gradient strip method was the least reliable form of AST, due to both the high percentage of discrepant MIC results and the percentage of category errors, especially VMEs for vancomycin and teicoplanin in the isolate with the VanB phenotype. These results are similar to those described in previous reports, in which gradient strips from Oxoid, Liofilchem and bioMérieux showed very low sensitivity (61%–63%) for detection of teicoplanin resistance in a collection of E. faecium isolates.22 The lack of reliability of MIC results using gradient strips could be related to several microbiological factors, such as inappropriate inoculum preparation or problems related to growth visualization. The limitation observed with gradient strips of vancomycin and teicoplanin would justify the use of more reliable alternative assays to avoid erroneous MIC results that could lead to false results of susceptibility (VMEs) and resistance (MEs).
The other non-automated AST method tested in the present study was an in-house broth microdilution method, which also produced a high percentage of VMEs for teicoplanin; nevertheless, these results should be interpreted with caution due to the low number of MIC results reported by those centers that used this AST method. As mentioned for gradient strips, vancomycin and teicoplanin MIC results obtained by in-house broth microdilution assays should be confirmed with other more reliable AST assays because of the clinical relevance of these microbiological results.
The semi-automated AST systems used in this study (MicroScan, Vitek 2 and Phoenix) produced a low and acceptable percentage of discrepant MICs, and a high percentage of VMEs, which were not related, as discussed below, with the use of the system, indicating that they are reliable AST method for VRE.
With respect to the breakpoints used for clinical categorization, those established by EUCAST were used twice as frequently as those of CLSI, which is in line with the gradual implementation of EUCAST guidelines in Europe.17,23,24 The errors obtained using EUCAST were slightly higher than those obtained with CLSI, which may be related to factors such as the “I” category (intermediate, as for EUCAST 2018), which was not defined by EUCAST for various antimicrobials (e.g. glycopeptides, linezolid, imipenem and levofloxacin), the wide MIC interval of the intermediate category) for some antimicrobials using CLSI (e.g. vancomycin), the lower breakpoints defined by EUCAST for some antimicrobials (e.g. teicoplanin), or the absence of breakpoints for some antimicrobials (e.g. daptomycin) with respect to CLSI (see below).
The type of antimicrobial was another factor contributing to MIC discrepancies and category errors. The highest percentages of discrepant MICs were obtained with imipenem and ampicillin, although the discrepancies were not associated with clinical category errors and should have no clinical impact. On the other hand, for teicoplanin, vancomycin and ciprofloxacin, although the discrepancies in MICs were low (<10%), they were associated with important errors (VMEs for teicoplanin and vancomycin, and MEs for ciprofloxacin) which could have significant clinical and epidemiological impact.
There was an unexpectedly very high percentage of VMEs obtained with teicoplanin, which was not related with obtaining discrepant MIC results with the AST system, but due to the interpretation of the VanB phenotype as teicoplanin-susceptible, in contrast to the EUCAST recommendations. The results of previous studies have highlighted the development of teicoplanin resistance in VanB E. faecium isolates, both in vitro and in vivo, leading to a high probability of therapeutic failure.14,25
The VMEs obtained with vancomycin, unlike those obtained with teicoplanin, were much lower and were related to discrepant MICs, particularly those generated by the use of MicroScan. All the vancomycin VMEs were observed in the CC-2 VanB isolate, and were true false susceptibilities associated with the reported vancomycin MICs, which were in the susceptible range (MIC of ≤4mg/l), in an isolate that was VR. It is important to highlight that false vancomycin susceptibility can represent a serious problem because this result is not routinely confirmed in the laboratory. Furthermore, failure to identify VRE isolates can lead to treatment failures that may be associated with increased mortality, poor clinical outcome, prolonged length of stay, or reinfections. Failure to recognize VRE may also have important epidemiological consequences, since vanA and vanB can be mobilized via plasmid transfer, contributing to the emergence of nosocomial outbreaks.
Ciprofloxacin was the only antimicrobial tested for which there were MEs (5.0%), but they may not have significant clinical implications. Ciprofloxacin is not frequently used as a first-line treatment for VRE infections, so that the clinical repercussions of reporting false ciprofloxacin resistance in VRE are scarce. Nevertheless, a report of ciprofloxacin as resistant may lead to the use of other antimicrobials with a broader spectrum and more toxic or with secondary effects, such as vancomycin or teicoplanin. The epidemiological impact of ciprofloxacin MEs is probably of low relevance since, in general, clinical ciprofloxacin resistance is not plasmid-encoded, and ciprofloxacin, is very infrequently used for the treatment of enterococcal infections.
The MEs with linezolid were acceptably low (1.1%), and their clinical implications should be similar to those reporting false ciprofloxacin resistance. From a microbiological point of view, it is important to confirm linezolid resistance with other AST systems, or by using a molecular assay to detect plasmid-encoded cfr, optrA and poxtA determinants, due to their potential clinical and epidemiological relevance.26 The epidemiological importance of linezolid resistance, in contrast to ciprofloxacin resistance, lies in the fact that acquired linezolid resistance may be plasmid-encoded (cfr, optrA and poxtA determinants), which could lead to the implementation of unnecessary control measures.
With respect to VR phenotype, most of the discrepant antimicrobial MICs and errors occurred with the VanB phenotype, which is consistent with the results of previous reports.22,27
An analysis of the potential factors that contribute to erroneous antimicrobial susceptibility results should be a priority for clinical laboratories. One way to address this problem is to participate in quality control programs, which can be very helpful for detecting potential laboratory problems and enabling corrective measures aimed at optimizing the processing and quality of the reports offered to clinicians to be established. This information is very useful for optimizing the best therapeutic strategies, improving the rational use of antimicrobials (reducing resistance rates), facilitating the control of nosocomial infections (by reducing the spread of MDR clones), and preventing outbreaks.28–30
The main limitation of the present study is its small sample size, which is typical of this kind of studies.
In conclusion, this study shows that the discrepancies in MIC values and error rates in the clinical categorization of VRE observed with some antimicrobials were associated with the AST system used, the application of EUCAST or CLSI breakpoints, the antimicrobial agent, and the interpretation of the VR phenotype. Most microbiology laboratories in Spain can reliably determine the antimicrobial susceptibility in VRE, although for teicoplanin there is a significant percentage of inadequate interpretations (false susceptibility), since VanB enterococci showing in vitro susceptibility to teicoplanin must be reported as resistant, with a warning indicating that evidence of resistance development during treatment with this antimicrobial has been reported.
FundingThis work was supported by the Instituto de Salud Carlos III, Subdirección General de Redes y Centros de Investigación Cooperativa, Ministerio de Economía y Competitividad, the Spanish Network for Research in Infectious Diseases (REIPI RD 16/0016)—co-financed by European Development Regional Fund ‘A way to achieve Europe (ERDF), and the Reference Laboratory, Program for the Prevention and Control of Healthcare-Associated Infections and Antimicrobial Stewardship in Andalucía (PIRASOA, Junta de Andalucía, Consejería de Salud Y Familias, Servicio Andaluz de Salud).
Conflict of interestThe authors declare that they have no conflict of interest.
We would like to thank the Reference Laboratory, Program for the Prevention and Control of Healthcare-Associated Infections and Antimicrobial Stewardship in Andalucía (PIRASOA, Servicio Andaluz de Salud), the Quality Control Programme (CCS) from the Spanish Society of Clinical Microbiology and Infectious Diseases (SEIMC), and the following 51 participating centers:
Hospital Universitario de Valme (Sevilla; J.L. García López), Hospital General de Gran Canaria Dr. Negrín (Las Palmas de Gran Canaria; A. Bordes Benítez), Hospital Costa del Sol (Málaga; N. Montiel), Hospital Universitario Miguel Servet (Zaragoza; A. Milagro Beamonte), Hospital de Cabueñes (Gijón; L. Otero Guerra), Hospital Doce de Octubre (Madrid; P. López Roa), Hospital Universitario Marqués de Valdecilla (Santander; J Calvo Montes), Hospital Santa Barbara de Soria (Soria; C. Aldea Mansilla), Hospital Clínico Universitario de Salamanca (Salamanca, J.L. Muñoz Bellido), Hospital Clínico Universitario de Valladolid (Valladolid; R. Ortiz de Lejarazu Leonardo), Hospital Universitario Rio Hortega (Valladolid; C. Ramos Sánchez), Hospital General Universitario de Albacete (Albacete; J. Lozano Serra), Hospital General Universitario de Ciudad Real (Ciudad Real; J.C. González Rodríguez), Hospital Virgen de la Salud (Toledo; C. Gómez Hernando), Hospital Sta. Creu i St. Pau (Barcelona; P. Coll Figa), Hospital Universitario de Bellvitge (Barcelona; M.A. Domínguez Luzón), Hospital Clínic (Barcelona; F. Marco), Hospital Virgen de Puerto (Plasencia; J.R. Muñoz del Rey), CHUVI Hospital Álvaro Cunqueiro (Vigo; R. Carballo Fernández), Complejo Hospitalario Universitario A CORUÑA (A Coruña; B. Fernandez Pérez), Hospital Infantil Niño Jesús (Madrid; M.M. Alonso Sanz), Hospital Universitario de la Princesa (Madrid; L. Cardeñoso Domingo), Hospital General Universitario Gregorio Marañón (Madrid; C. Sánchez), Hospital Clínico Universitario San Carlos (Madrid; J. Prieto Prieto), Hospital Universitario Puerta de Hierro Majadahonda (Madrid; F. Portero), Complejo Hospitalario de Navarra (Pamplona; X. Beristain Rementeria), Clínica Universidad de Navarra (Pamplona; J. Leiva León), Hospital Universitario de Álava (Vitoria; A. Canut Blasco), Hospital Universitario Donostia (Donosti-San Sebastián; C. Gustavo Cilla), Hospital de Cruces (Barakaldo; I. Perales Palacios), Hospital de Galdakao (Galdakao; M.J. López de Goicoechea S. Román), Hospital Marina Baixa (Villajoyosa; C. Martínez Peinado), Consorcio Hospital General Universitario de Valencia (Valencia; N. Tormo), Hospital Universitario y Politécnico La Fe (Valencia; J.L. López Hontangas), Hospital General Universitario de Elche (Elche; N. Gonzalo Jiménez), Hospital Universitario Puerta del Mar (Cádiz; M.A. Rodríguez Iglesias), Hospital Nª Sra de la Candelaria (Santa Cruz de Tenerife; A. Sampere Martínez), Hospital Universitario Virgen de la Victoria (Málaga; E. Clavijo Frutos), Hospital Severo Ochoa (Leganés; S. Quevedo), Complejo Hospitalario de Pontevedra (Pontevedra; A. Pallares González), Hospital Virgen de las Nieves (Granada; J.M. Navarro Marí), Hospital Universitario Reina Sofía (Córdoba; I. Gracia Ahufinger), Complejo Asistencial Universitario de León-Sacyl (León; M.I. Fernández Natal), Hospital Universitario La Paz (Madrid; M.R. Gómez Gil), Hospital Universitario Son Espases (Palma de Mallorca; J.L. Pérez Sáenz), Hospital Ramón y Cajal (Madrid; R. Cantón), Hospital Virgen de Altagracia (Manzanares; A. Sánchez-Maroto Lozano), Hospital Universitario Fundación Alcorcón (Alcorcón; A. Delgado), Hospital Universitario Vall d́Hebron (Barcelona; L. Goterris), Hospital Clínico Universitario Virgen de la Arrixaca (El Palmar; G. Yague Guirao), Hospital Universitario Virgen del Rocío (Sevilla; J. Aznar Martín).