NDM-1 carbapenemase is spreading rapidly all over the world, but this metallo-beta-lactamase has just been detected for the first time in an Acinetobacter baumannii (Ab) isolate of the ST85 clone in Spain. The aim of this study was to characterize a NDM-1-producing carbapenem-resistant A. baumannii (CR-Ab) isolate submitted to the Andalusian PIRASOA [infection prevention program] referral laboratory.
MethodsCarbapenemases were detected by PCR and Sanger DNA sequencing. Whole genome sequencing was performed by NGS (Miseq, Illumina). Resistance genes were identified with RESfinder, while MLSTfinder was used for sequence typing (ST). The genetic location of blaNDM-1 was determined by nuclease S-1/PFGE/hybridization with specific probe.
ResultsThe isolate was susceptible to amikacin and tigecycline and belonged to the ST85 clone. blaOXA-94 and blaNDM-1 were identified by PCR and Sanger DNA sequencing, respectively. The resistance genes aadB, blaADC-25, blaNDM-1, blaOXA-94, msr(E), mph(E) and floR,sul2 were identified by NGS. The chromosome of the isolate contained a defective Tn125 transposon with blaNDM-1 flanked by the insertion sequences ISAbA125 and ISAba14. The blaNDM-1 gene was only detected in the chromosomal DNA.
ConclusionThis is the first time that blaNDM-1 has been detected and characterized in a blaOXA-94-producing CR-Ab isolate belonging to the ST85 clone in Spain.
La carbapenemasa NDM-1 se está diseminando rápidamente por todo el mundo, pero esta metalo-beta-lactamasa se detecta por primera vez en un aislado de A. baumannii del clon ST85 procedente de España. El objetivo de este estudio es caracterizar un aislado de A. baumannii resistente a carbapenémicos productor de NDM-1 remitido al laboratorio de referencia PIRASOA de Andalucía.
MétodosLa detección de carbapenemasas se realizó mediante PCR y secuenciación de ADN Sanger. La secuenciación del genoma completo se realizó mediante NGS (MiSeq, Illumina). La detección de genes de resistencia y el secuenciotipo (ST) se obtuvo mediante ResFinder y MLSTFinder, respectivamente. La localización de blaNDM-1 se determinó utilizando el método de la nucleasa S1/PFGE/hibridación con sonda específica.
ResultadosEl aislado era sensible a amikacina y tigeciclina, y pertenecía al clon ST85. Se identificaron las variantes blaOXA-94 y blaNDM-1, respectivamente, mediante PCR y secuenciación Sanger. Mediante secuenciación masiva se detectaron los genes de resistencia aadB, blaADC-25, blaNDM-1, blaOXA-94, msr(E), mph(E), floR y sul2. El aislado contenía en su cromosoma un transposón defectivo de tipo Tn125 con blaNDM-1 flanqueado por las secuencias de inserción ISAbA125 y ISAba14. El gen blaNDM-1 solo se detectó en el ADN cromosómico.
ConclusiónEn este estudio se detecta y se caracteriza por primera vez en España blaNDM-1 en un aislado de A. baumannii resistente a carbapenémicos productor de blaOXA-94 y perteneciente al clon ST85.
Acinetobacter baumannii is a successful pathogen characterized by its ability to acquire multidrug resistance (MDR) and to cause nosocomial outbreaks. Resistance to carbapenems in A. baumannii has increased in recent years and has been related to the production of carbapenemases, alterations in porins, overexpression of efflux pumps and altered penicillin-binding proteins.1 The main mechanism of resistance to carbapenems in A. baumannii is the acquisition of genes coding for carbapenem-hydrolyzing class D beta-lactamase (CHDL), also known as oxacillinases (blaOXA-23, blaOXA-24/40, blaOXA-58, blaOXA-143 and blaOXA-235).2 The role of the naturally occurring chromosomic blaOXA-51-like oxacillinases (i.e. blaOXA-51, blaOXA-66, blaOXA-69, blaOXA-71, blaOXA-94) in the resistance to carbapenems is unclear. Class B beta-lactamases (metallo-beta-lactamases; MBL) (i.e. blaVIM-like) are generally much less frequent than oxacillinases in carbapenem-resistant A. baumannii. Nevertheless, MBLs tend to hydrolyze carbapenems more efficiently than oxacillinases and have a broader spectrum of hydrolytic activity (including expanded-spectrum cephalosporins and aztreonam).2,3 Some of these MBLs, like the New Delhi metallo-β-lactamase-1 (NDM-1), are rapidly spreading worldwide in gramnegative bacteria, which represent a serious problem from a clinical and an epidemiological point of view.4,5 In A. baumannii, the gene encoding NDM-1 has been detected on the chromosome and, less frequently, in plasmids from isolates from different countries around the world.6 Since this is the first time that NDM-1-producing A. baumannii has been detected in Spain, we decided to (i) characterize this isolate; (ii) determine the genetic localization (chromosomal or plasmidic) of blaNDM-1 and (iii) analyze its genetic environment.
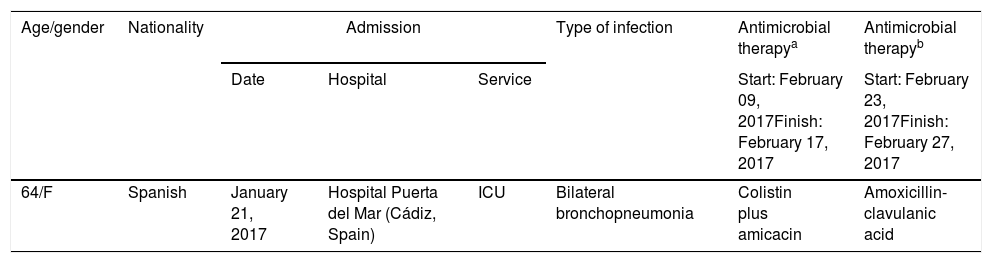
Material and methodsBacterial isolateA carbapenem-resistant A. baumannii isolate obtained from a rectal swab (Table 1 shows the most relevant clinical features) from a patient in April 18, 2017 was submitted to the Andalusian reference laboratory for multidrug-resistant nosocomial pathogens (PIRASOA program, Seville, Spain) for characterization (see below).
Relevant clinical features.
| Age/gender | Nationality | Admission | Type of infection | Antimicrobial therapya | Antimicrobial therapyb | ||
|---|---|---|---|---|---|---|---|
| Date | Hospital | Service | Start: February 09, 2017Finish: February 17, 2017 | Start: February 23, 2017Finish: February 27, 2017 | |||
| 64/F | Spanish | January 21, 2017 | Hospital Puerta del Mar (Cádiz, Spain) | ICU | Bilateral bronchopneumonia | Colistin plus amicacin | Amoxicillin-clavulanic acid |
Identification of genomic species of Acinetobacter was performed by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry, MALDI-TOF MS (MALDI Biotyper CA system; Bruker Daltonics, Madrid, Spain). Detection of blaOXA-51 was performed by PCR,7 whole genome sequencing (WGS) and the SpeciesFinder 1.2 tool (https://cge.cbs.dtu.dk/services/SpeciesFinder/).
Antimicrobial susceptibility testing was performed with the Microscan Neg MIC Panel, Type 44 (Beckman Coulter, Inc, Madrid, Spain). Imipenem and meropenem susceptibility testing was performed by Etest (LioChem Inc, Madrid, Spain) and disk diffusion (Oxoid, Madrid, Spain) in Mueller-Hinton agar plates (Oxoid). Susceptibility to colistin was tested by microdilution using the UMIC kit (Biocentric; Bandol, France).
The isolate was genotyped by PFGE using the restriction enzyme ApaI,8 multilocus sequence typing (MLST) was performed according to the Institut Pasteur scheme (https://pubmlst.org/abaumannii/) and using WGS (see below) and the MLST 1.8 tool (https://cge.cbs.dtu.dk/services/MLST/).
Detection of antimicrobial resistance genesPhenotypic detection of carbapenemases was performed with the combination disk test (Rosco, Madrid, Spain) according to EUCAST guidelines.9
Detection of genes coding for carbapenemases was carried out by PCR using specific primers for class A (blaKPC), B (blaIMP, blaVIM, blaNDM) and D (blaOXA-23, blaOXA-24/40, blaOXA-58, blaOXA-48) carbapenemases.7,10 Allelic variants positive by PCR were determined by Sanger sequencing (Macrogen, Madrid, Spain). Other genes associated with resistance to antimicrobials were also identified using WGS (see below) and the ResFinder 3.0 tool (https://cge.cbs.dtu.dk/services/ResFinder/).
Conjugation and transformation assaysConjugation experiments were carried out in Luria–Bertani broth (LB, Oxoid) with sodium azide-resistant Escherichia coli J53 and Acinetobacter baylyi (MIC of rifampicin 16mg/L) as the recipients. Transconjugants were selected by plating onto LB agar plates (Oxoid) supplemented with 100mg/L of sodium azide and 0.125mg/L of ertapenem (Sigma, Madrid, Spain) using E. coli J53 as recipient, and 8mg/L of rifampicin (Sigma, Madrid, Spain) and ertapenem, respectively, using A. baylyi as recipient.
Plasmid DNA was extracted by the Kieser method, electroporated into E. coli DH10B and plated on MacConkey agar (Becton Dickinson, Madrid, Spain) supplemented with ertapenem at 0.125mg/L.11
Whole genome sequencingThe genome of the isolate was sequenced with next-generation sequencing (NGS) using the Illumina MiSeq system. The sample library was prepared with the Nextera XT DNA library preparation kit (Illumina, San Diego, CA, USA). DNA sequencing was carried out with the MiSeq Reagent Kit V3 (600 cycles) and the Illumina MiSeq sequencer (2×300 paired end reads). The reads were quality filtered. De novo assembly and gene annotation was performed using the CLC Genomics Workbench v10 (Qiagen) and the RAST server (http://rast.nmpdr.org/), respectively. Characterization of the resistome was carried out using the ResFinder server (http://cge.cbs.dtu.dk/services/ResFinder-3.0).
Plasmid analysis and Southern hybridizationPlasmid size was determined by electrophoresis using a 0.7% agarose gel and E. coli 50192 harboring 154, 66, 48 and 7kb plasmids as size markers. The genetic localization of blaNDM-1 was determined by Southern blotting. Genomic DNA was digested with nuclease S1 (Roche, Madrid, Spain), separated by PFGE electrophoresis, transferred to a positively charged nylon membrane (Amersham Hybond-N+, Madrid, Spain) and hybridized with a digoxigenin-labeled blaNDM-1 probe.
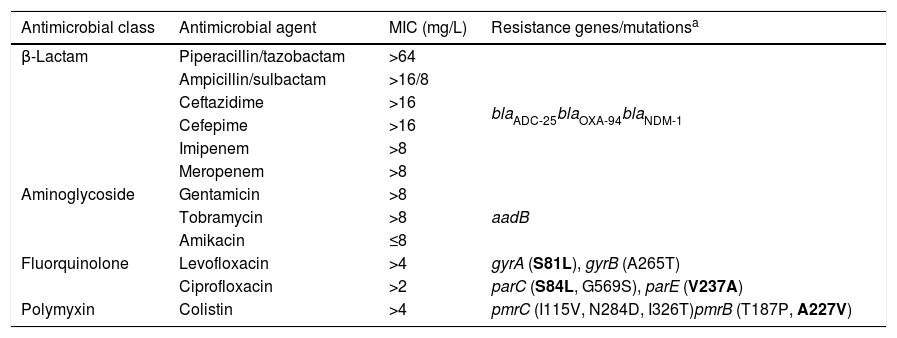
ResultsThe isolate (Ab-NDM-1) was identified by MALDI-TOF MS and the SpeciesFinder 1.2 tool as A. baumannii, and was positive for blaOXA-51 by PCR. MICs obtained by microdilution using Type 44 panels are shown in Table 2. Etest MICs were >32mg/L for imipenem and meropenem, and 1mg/L for tigecycline. The isolate was susceptible only to amikacin according to EUCAST and CLSI breakpoints of 2018.12,13 The MIC of colistin was >64mg/L using the UMIC kit.
MICs and resistome of the NDM-1 producing A. baumannii isolate.
| Antimicrobial class | Antimicrobial agent | MIC (mg/L) | Resistance genes/mutationsa |
|---|---|---|---|
| β-Lactam | Piperacillin/tazobactam | >64 | blaADC-25blaOXA-94blaNDM-1 |
| Ampicillin/sulbactam | >16/8 | ||
| Ceftazidime | >16 | ||
| Cefepime | >16 | ||
| Imipenem | >8 | ||
| Meropenem | >8 | ||
| Aminoglycoside | Gentamicin | >8 | aadB |
| Tobramycin | >8 | ||
| Amikacin | ≤8 | ||
| Fluorquinolone | Levofloxacin | >4 | gyrA (S81L), gyrB (A265T) |
| Ciprofloxacin | >2 | parC (S84L, G569S), parE (V237A) | |
| Polymyxin | Colistin | >4 | pmrC (I115V, N284D, I326T)pmrB (T187P, A227V) |
The isolate was assigned to ST85 by MLST and the MLSTFinder 1.8 tool, and was clonally unrelated (more than 5 band differences) to previous pulsotypes of carbapenem-resistant A. baumannii in the Andalusian database of the PIRASOA program reference laboratory. Synergy was observed between the meropenem disks and the dipicolinic acid disks. blaOXA-51-type and blaNDM-type genes were detected by endpoint PCR, and were identified as blaOXA-94 and blaNDM-1, respectively, by Sanger sequencing.
After trimming, 578,845 reads were obtained by WGS. The average length of these reads was 260.3bp and GC content was 39.5%. After assembly, the draft genome consisted of 316 contigs, with mean length of 22,375bp and mean sequencing coverage of 53, yielding a total read length of 3,950,783bp. After genome annotation using Rapid Annotations using Subsystems Technology (RAST) server (http://rast.theseed.org/FIG/rast.cgi), a total of 3703 coding sequences and 78 RNAs were obtained.
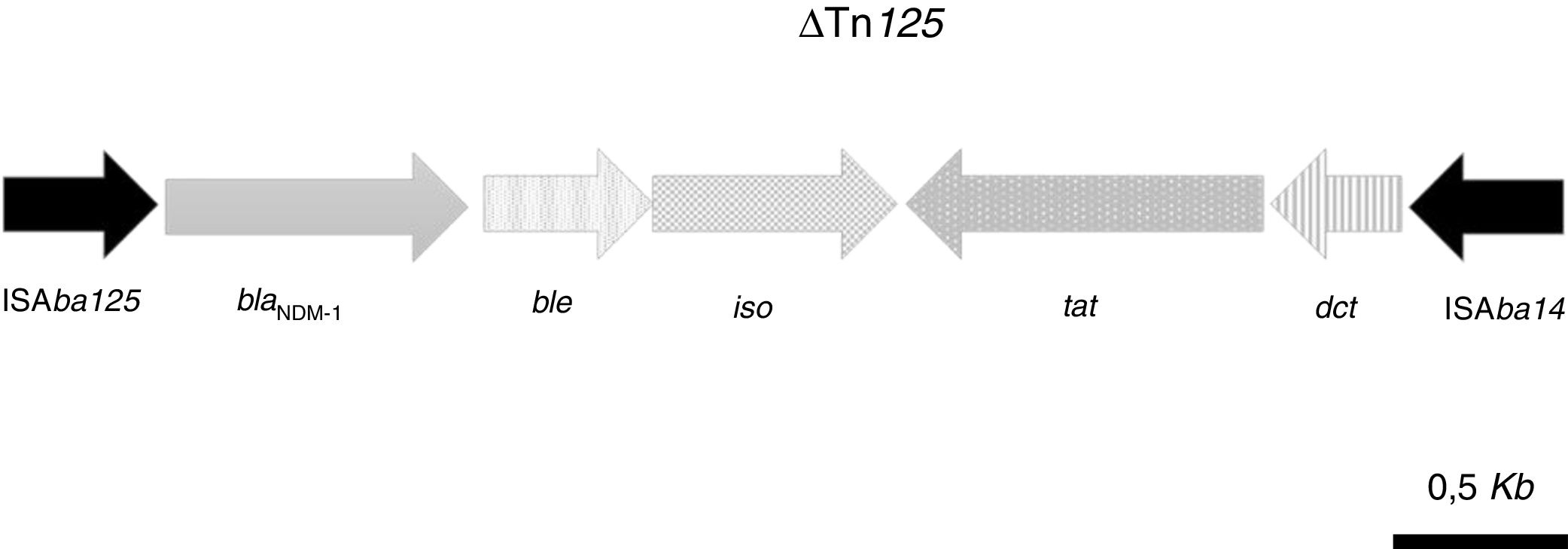
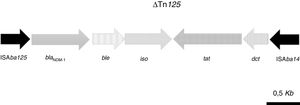
The acquired antimicrobial resistance genes identified in the assembled contigs with the ResFinder 1.2 tool were associated with resistance to aminoglycosides (aadB, which confers resistance to gentamicin and tobramycin), beta-lactams (chromosomal cephalosporinase blaADC-25, class B carbapenemase blaNDM-1 and the OXA-51-type CHDL, blaOXA-94), macrolides, lincosamides and streptogramins [msr(E), mph(E)], phenicols (floR) and sulfonamide (sul2). The resistome for the most relevant group of antimicrobials is shown in Table 2. WGS revealed that blaNDM-1 was located on a defective transposon (ΔTn125) flanked by the insertion sequences ISAba125 and ISAba14 inserted in the upstream and downstream regions, respectively (Fig. 1).
Schematic representation of the genetic environment of blaNDM-1: ble gene, encoding the bleomycin resistance protein; iso gene, encoding a putative phosphoribosylanthranilate isomerase; tat gene, encoding oxidoreductase DsbD superfamily protein; dct, gene, encoding divalent cation tolerance protein.
With respect to chromosomal mechanisms related to carbapenem resistance, mutations in the porin genes, carO and oprD-like, were not detected, whereas several point mutations (V90I, H127K, S168G, T173S, D183N, S236T, S243E and V270L) were observed in the omp33-36 gene. No mutations were detected in genes coding for the two-component regulatory system adeR-adeS, which regulates the expression of genes encoding the efflux pump adeABC.
With respect to colistin resistance, acquired resistance genes (mcr-1) or mutations in the chromosomal genes pmrA, lpxA, lpxC and lpxD were not detected. Table 2 shows the aminoacid changes detected in pmrB and pmrC.
The isolate carried three different plasmids, which were ∼150kb, ∼37kb and ∼6kb in size, respectively. No transconjugants or transformants producing NDM were obtained. The blaNDM-1 probe hybridized to chromosomal DNA, but not to any of the three plasmids observed.
Nucleotide sequence accession numberThis draft genome project has been deposited registered at GenBank (accession number QBY00000000.1) with the BioProject ID PRJNA449628 (http://www.ncbi.nlm.nih.gov/bioproject/449628).
DiscussionIn countries all over the world, the acquired carbapenemase NDM-1 is spreading efficiently in several gramnegative microorganisms in nosocomial and community-acquired infections, which poses a major challenge for the treatment and control of healthcare-associated infections.4,5,14,15 In A. baumannii, this MBL has been detected in carbapenem-resistant isolates in different countries of Europe (Switzerland, Slovenia, Germany, France, Belgium, Czech Republic, Turkey), Latin America (Colombia), Africa (Algeria, Ethiopia, Syria,) and Asia (India).15–21 In Spain, NDM-1-producing A. baumannii has not been reported before.
Isolate Ab-NDM-1 showed cross-resistance with other antimicrobial families (quinolones and some aminoglycosides) consistent with other NDM-1-producing isolates in other countries. This reduces the therapeutic options for infection to certain toxic antimicrobials such as colistin.14 Although this isolate lacked some of the most frequently described CHDL genes in A. baumannii (OXA-23-like, OXA-24/40-like, OXA-58-like), carbapenem resistance is fully explained by the acquisition of blaNDM-1. Nevertheless, additional carbapenem resistance mechanisms, probably related to the Omp 33–36 porin, may be implicated, as described in A. baumannii isolates in other studies.22–24 Although some of the amino acid changes observed in Omp 33–36 of Ab-NDM-1 could be associated with decreased susceptibility to carbapenems, additional studies with isogenic mutants are required to understand their role in carbapenem resistance.
The absence of mutations in the regulatory genes adeS and adeR suggests that the efflux pump adeABC, implicated in A. baumannii resistance to some antimicrobials and biocides, is not overexpressed in Ab-NDM-1, which is in agreement with the absence of resistance to tigecycline.25,26
Isolate Ab-NDM-1 also showed resistance to colistin, which was associated with some aminoacid changes in pmrC and particularly in pmrB (i.e. A227V). The implication of other less frequent or undescribed mechanisms of colistin resistance related to the synthesis of lipid A cannot be ruled out.27
In Enterobacteriaceae, blaNDM-1 is frequently plasmid encoded, whereas it is more frequently found on the chromosome in A. baumannii, although it has also been detected on plasmids.6,15,16 Similar genetic environments have been detected harboring blaNDM-1 in Enterobacteriaceae and A. baumannii, indicating horizontal transmission (plasmid transfer) of this carbapenemase, probably from Enterobacteriaceae to A. baumannii.28
The carbapenemase blaNDM-1 was detected on the chromosome of Ab-NDM-1, but not in any of the three plasmids observed. This finding suggests that the clone must have been introduced into the Hospital Puerta del Mar by clonal dissemination, rather than by plasmid transfer from a NDM-1-producing gramnegative isolate not previously detected in this center. Another possible hypothesis that was not investigated in this study is phage-mediated transfer of blaNDM-1, as has been described among isolates of A. baumannii.6
Tn125-like transposons are the most frequent genetic platforms for mobilization of blaNDM-1.15 In Ab-NDM-1 isolates, the blaNDM-1 carbapenemase is associated with a truncated transposon (ΔTn125) containing a genetic environment similar or identical to those previously described in other carbapenem-resistant NDM-1- and OXA-94-producing A. baumannii isolates, particularly those belonging to the ST85 clone previously detected in some countries (France, Algeria, Turkey, Syria, Tunisia) (19–22,30).18–21,29 In Spain, the A. baumannii ST85 clone has only been detected in OXA-23-producing carbapenem-resistant isolates causing nosocomial outbreaks.30
Tn125-like transposons have also been detected in A. baumannii clones other than ST85, such as clone ST25 (in Slovenia, Germany, Lebanon), which is not related to CC92/CC2, but included in a different CC together with the ST6 clone belonging to international clonal lineage I (in Switzerland).18–21
The patient with Ab-NDM-1 made no reference to any history of travel to other countries where NDM-1-producing bacteria are prevalent, so that the source of this Ab-NDM-1 isolate in the Hospital Puerta del Mar remains unclear. At the same time, contact between this patient and other patients or healthcare personnel colonized with NDM-1-producing gramnegative microorganisms cannot be ruled either.
This study has some limitations. One of them is the lack of epidemiological and clinical data about other patients admitted to the same center, which makes difficult to investigate potential reservoirs and transmission routes, or the country of origin of the Ab-NDM-1 isolate.
In conclusion, to the best of our knowledge, this is the first time that blaNDM-1 has been detected in blaOXA-94-producing carbapenem-resistant A. baumannii belonging to the ST85 clone in Spain.
FundingThis work was partially funded by a grant (PI11/02046) from the Instituto de Salud Carlos III, Subdirección General de Redes y Centros de Investigación Cooperativa, Ministerio de Economía, Industria y Competitividad.
Conflict of interestNone to declare.
This work was presented in part at the 2018 ECCMID in Madrid (Poster 1410).
This work was supported by the Plan Nacional de I+D+i 2013–2016 and the Instituto de Salud Carlos III, Subdirección General de Redes y Centros de Investigación Cooperativa, Ministerio de Economía, Industria y Competitividad, Spanish Network for Research in Infectious Diseases (REIPI; RD16/0016/0001)-co-financed by European Development Regional Fund “A way to achieve Europe”, Operative program Intelligent Growth 2014–2020.
We would like also to thanks to María del Mar Tomás Carmona (Hospital de A Coruña, Spain) for her kindness in giving us a strain of Acinetobacter baylyi.