Livestock are known reservoirs of methicillin-resistant Staphylococcus aureus (MRSA) and this constitutes an important public health issue. The prevalence of nasal MRSA carriers in swine housed indoors in Galicia, Spain, was studied.
Methods197 samples from swine aged three, eight, 12, 16 and 24 weeks, and from adult pigs, were obtained from four farms. The cleaning procedures implemented to clean the barns and antimicrobial consumption were analyzed. Antimicrobial susceptibility and antimicrobial resistance genes were studied. PFGE, spa typing and MLST were used to classify the isolates. SCCmec, agr and pvl were analyzed.
ResultsMRSA prevalence was 12.7%. Swine younger than 16 weeks had a higher colonization rate; 22.9% vs 3.5% (OR, 8.16; 95% CI, 2.47–29.79; p<0.01). The only farm found to be MRSA-free used disinfectants as part of its cleaning procedure. All MRSA were tetracycline-resistant (identifying the tetK and tetM genes), 80% were resistant to erythromycin and clindamycin and 16% were only clindamycin-resistant. The ermC and vgaA genes were identified in these two phenotypes. A single genotype (PFGE type A) and ST398 – spa t011 (84%) and t1451 (16%) were identified. SCCmec type V and agrI were identified in all isolates, and all were pvl-negative.
ConclusionA correlation between swine age and MRSA colonization was observed. Appropriate cleaning procedures could have an impact on MRSA colonization in farming. Resistance to antibiotics used in human health was identified. Clinicians should be aware if their patients have come into contact with farm animals.
Los animales de granja son reservorios de Staphylococcus aureus resistente a la meticilina (SARM), y constituyen un problema de salud pública. Se estudia la prevalencia de portadores nasales de SARM en cerdos estabulados en Galicia, España.
MétodosEn 4 explotaciones se obtuvieron 197 muestras de cerdos con edades en semanas de 3, 8, 12, 16, 24 y adultos. Se analizaron los métodos empleados para limpiar los establos y el consumo de antimicrobianos. Se estudió la resistencia a antimicrobianos, y los genes involucrados en esta. Los aislamientos fueron clasificados mediante PFGE, spa y MLST. Se analizaron SCCmec, agr y pvl.
ResultadosLa prevalencia de SARM fue del 12,7%. Los cerdos de <16 semanas presentaron las frecuencias de colonización más elevadas 22,9 vs. 3,5% (OR: 8,16; IC 95%: 2,47-29,79; p<0,01). En la única explotación libre de SARM se empleaban desinfectantes en la limpieza. Todos los SARM fueron resistentes a tetraciclina identificándose los genes tetK y tetM, el 80% fueron resistentes a eritromicina y clindamicina, y el 16% fueron únicamente resistentes a clindamicina. Se identificaron los genes ermC y vgaA en estos 2 fenotipos. Se identificó un único genotipo (PFGE-A) y ST398, siendo spa t011 (84%) y t1451 (16%). En todos los aislamientos se identificó SCCmec V y agrI, siendo estos pvl negativos.
ConclusionesSe observó la asociación entre edad y colonización SARM. La limpieza adecuada podría modificar la colonización por SARM. Se detectaron resistencias a antibióticos empleados en humanos. Los médicos deberían conocer si los pacientes tienen contacto con animales de granja.
In 2005 a methicillin-resistant Staphylococcus aureus (MRSA) strain associated with pig farming was identified in France and The Netherlands.1,2 The MRSA strain belonged to sequence type (ST) 398 clone which has also been further identified in animals and humans worldwide.3–6 Indeed, 20% of all MRSA identified in humans in The Netherlands belonged to the ST398, documenting the importance of considering livestock and other animals when studying the epidemiology of MRSA.7
Spain is the fourth porcine producer in the world and the second in Europe behind Germany.8 The current information about MRSA colonization in live swine in Spain is scarce. Porrero et al.9 studied the MRSA epidemiology in free living Iberian pigs and Reynaga et al.3 studied MRSA colonization in farm pigs. Other Spanish studies were done in slaughter pigs10 and food samples.11 Nevertheless, pigs could have become MRSA colonized during transportation from farm to slaughterhouse12 not representing the real farm prevalence.
In 2006, ST398 MRSA was first isolated from patients with different infections in our hospital.13 We concluded that the ST398 clone was part of the MRSA population in Galician region (Spain), hypothesizing that pigs could be a potential reservoir of MRSA from a public health perspective.
The aim of this study was to determine the prevalence of nasal MRSA carriage in live swine hosted in indoor production systems in Galician region, Spain. The isolates were genetically classified and the antimicrobial resistance was studied. The cleaning procedures used in each of the studied farms were questioned.
Materials and methodsFarms descriptionThe four sampled farms were located in an area of 80km2 in Galicia (Spain). From March to April 2010 we collected a total of 197 nasal swabs from two farrow-to-rearing farms (farms A and B, pigs were up to 8 weeks), a rearing farm (farm C; 8–24 weeks, at the moment of the study the pigs were 16 weeks), and a farrow-to-finish farm (farm D; 90 sows, 140 piglets 3 weeks old, 170 grower pigs 12–24 weeks old). The sample comprised 28 piglets 3 weeks old, 45 weaned piglets 8 weeks old, 10 grower pigs 12 weeks old, 75 grower pigs 16 weeks old, 25 finisher pigs 24 weeks old, and 14 adult breeding sows (Table 1). The farms were questioned about the cleaning procedures and the antimicrobial consumption. The origin of the swine was also inquired. The permissions for this study were obtained from the farm owners allowing us to sample a previously defined number of animals. For the development of this study the European Directive 2010/63/EU was fulfilled, which is transposed to Spanish national legislation by means of Real Decreto (RD) 53/2013 for the protection of animals for experimentation and other scientific purposes.
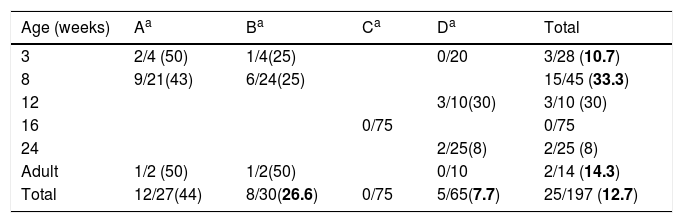
Swine age and methicillin-resistant Staphylococcus aureus (MRSA) isolated by farm. Farm denomination and MRSA/total S. aureus (%).
A total of 197 nasal swine samples were collected using sterile swabs and inserted in Stuart's medium at 4°C for transportation. The two nares were sampled with the same swab. The number of swine sampled represented 6.8% of the total pigs hosted in the farms at the moment of the study. Samples were not pooled, so they were inoculated one by one into 2mL enrichment broth containing 10g tryptone/L, 75g mannitol/L and 2.5g yeast extract/L.4 After 24h of incubation at 35°C, a loopful of broth was inoculated onto selective MRSA agar plate (ChromID MRSA, bioMérieux) and colistin-nalidixic acid agar with 5% lamb blood (CNA) (bioMérieux). The plates were incubated for 24–48h at 35°C and examined for MRSA. Only one MRSA isolate was selected for further analysis. Isolates were confirmed to be S. aureus by the catalase test, the tube coagulase test and biochemically identified using the VITEK 2 GP card (bioMérieux). Methicillin resistance was confirmed by testing for the presence of penicillin binding protein 2 (PBP2’) (MRSA latex agglutination test, Oxoid).
Pulsed field gel electrophoresis (PFGE) analysisWhole DNA from each MRSA isolate was analyzed by SmaI, Cfr9I and EagI, as previously described using the CHEF DR-III System (Bio-Rad).4,14,15S. aureus NCTC 8325 was included as reference strain. Genetic similarity between profiles was determined by computer analysis of the banding patterns using the InfoQuest FP software (Bio-Rad). Percent similarities were identified on a dendrogram derived from the unweighted pair group method using arithmetic averages and based on Dice coefficients. Band position tolerance and optimization were set at 1.0 and 0.5%, respectively. A similarity coefficient of 80% was selected to define the pulsed-field type clusters. Every band difference within a PFGE type resulted in adding a numerical order to the pulsed field cluster.16
Spa typing, multilocus sequence typing (MLST), staphylococcal chromosome cassette (SCCmec) typing, and agr typingDNA was extracted using the RTP-bacteria minikit (STRATEK Molecular). All isolates were analyzed by spa typing and MLST.17,18 MRSA isolates were SCCmec typed.19 The agr type was also identified.20 The presence of the virulence pvl gene was determined by PCR.21
Antimicrobial susceptibility testing and detection of antibiotic resistance genesAll MRSA were tested for antimicrobial susceptibility by the VITEK 2 AST-P588 card (bioMérieux, France). Susceptibility to tetracycline and daptomycin was studied using Etest on Mueller-Hinton agar (bioMérieux, France). The results were interpreted following the EUCAST recommendations.22S. aureus ATCC 29213 was used for quality assurance. Antimicrobial resistance genes were detected by PCR amplification as previously described.23–25
Data analysisChi-square test was used to analyze the statistical significance. A significance level of 0.05 was used in the analysis.
ResultsDetection of MRSA in the studied samples, cleaning procedures used, pig's origin, and antimicrobial consumptionNasal swabs were taken from 197 swine hosted in four farms representing 6 different age groups (Table 1). The overall prevalence was 12.7% (n=25). Swine younger than 16 weeks old had higher odds of MRSA colonization 22.9 vs 3.5% (OR, 8.16; 95% CI, 2.47–29.79; p<0.01, Chi square test) when compared to swine older than 16 weeks. No MRSA was isolated in the farm C where all the pigs were 16 weeks old.
The cleaning procedure used in farm C included farm emptying, wash using high-pressure water, disinfection using 250-PPM bleach solution pulverization, washed again with high-pressure water, and finally painted with quicklime. This procedure was repeated three times per year. The cleaning procedures in the other three farms comprised just cleaning with high-pressure water.
The farms informed that swine came solely from other local farms in Galicia. As well, they denied any antimicrobial use or consumption in the farms.
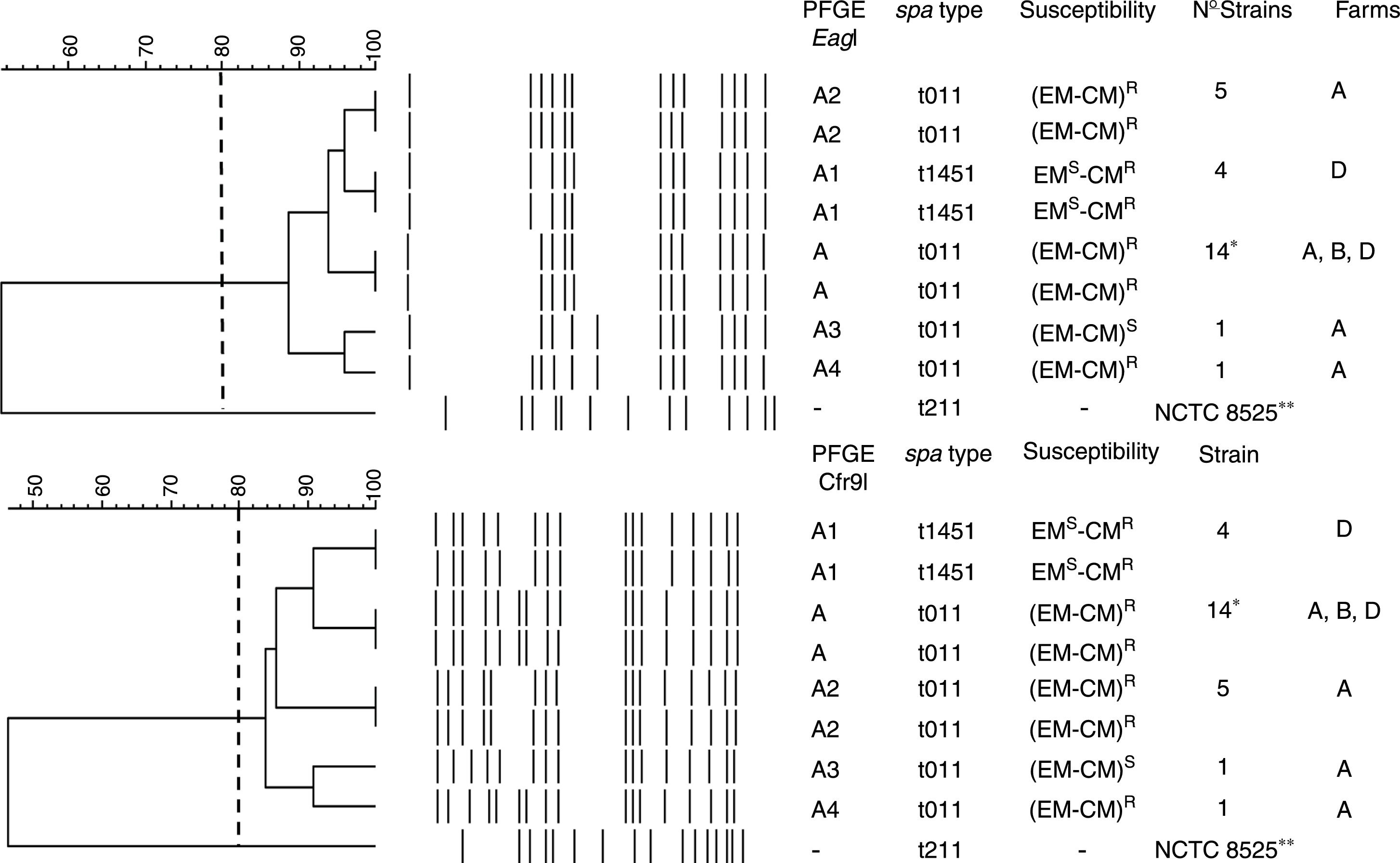
PFGE, spa, MLST, and SCCmec typing of MRSA isolatesTwenty-five MRSA isolates were subjected to PFGE. The isolates were non typeable when the DNA was digested with SmaI but they were typeable with EagI and Cfr9I. All isolates were closely related, Dice coefficient of similarity ≥ 0.84, being identified one PFGE cluster and four PFGE subtypes (Fig. 1).
Dendrogram showing the PFGE relatedness using EagI and Cfr9I. The Dice coefficient of similarity (D) was 0.84. One cluster, A, was detected and four PFGE subtypes were identified (A1, A2, A3, and A4). The spa types, susceptibility to erythromycin (EM) and clindamycin (CM), number of isolates per each PFGE and spa types, and the farms where each PFGE and spa where isolated are shown. * Farm A, 12 strains; farm B, 8 strains; and farm D, 5 strains. R and S mean resistant and susceptible respectively. -, no susceptibility studied, **NCTC 8325, its chromosomal DNA was digested with SmaI.
The spa-type t011 was identified in 21 isolates (84%). The spa-type t1451 was identified in 4 isolates (16%) (Fig. 1). All isolates belonged to the ST398 clone.
All isolates were SCCmec type V, agrI type and pvl-negative.
Antimicrobial susceptibility and resistance genesAll isolates were methicillin and tetracycline resistant. They were susceptible to trimethoprim-sulfamethoxazole, gentamicin, tobramicin, levofloxacin, linezolid, daptomycin, vancomycin, teicoplanin, rifampicin, mupirocin, fusidic acid, fosfomycin, nitrofurantoin, and tigecycline. Twenty strains (80%) were resistant to erythromycin (EM) and clindamycin (CM) and this resistance was constitutive. Four strains (16%) were resistant to CM but susceptible to EM, and one strain was EM and CM susceptible (4%) (Fig. 1).
The tetK and tetM genes were detected in all isolates, the tetL gene was not identified. The ermC gene was detected in all EMR-CMR isolates. The ermA and B genes were not detected in any isolate. The vgaA gene was identified in four EMS-CMR isolates, all belonging to spa-type t1451.
DiscussionGalicia is one of the western regions in the European Union. The overall prevalence of MRSA carriage in pigs (12.7%) was lower to the one reported by other series.3–5,26 Remarkably, no MRSA was identified in farm C were all pigs were 16 weeks old. In this production, pigs were hosted for four months before being delivered for consumption. Once the facility was totally emptied, an intensive cleaning procedure was always performed with cheap products like water, bleach and quicklime painting. As has been suggested by others, the presence of MRSA among pigs is related to the introduction of MRSA positive animals in a MRSA negative scenario, also the herd size could influence in the MRSA prevalence.5,27 The origin of the pigs was questioned, and it was verified that local farms were the only pig suppliers. That makes us to think that MRSA is a usual colonizer among swine in Galicia. The different cleaning procedures and the pig's age could be an important factor in the absence of swine's MRSA colonization in farm C. Moreover, MRSA has been identified in dust samples obtained from farms and from the trucks used for pig transportation. Therefore, these studies underline the role of the environment as a source of MRSA in the pig production chain.6,12,27
In this work the rate of colonization by SARM was higher in younger pigs, as previously showed by Smith TC et al.4
According to other national studies,3,9 the spa-type t011 was the most prevalent (84%). In this study, t1451 was isolated from 4 pigs only in farm D and four PFGE subtypes in the t011 spa-type were identified (Fig. 1). Therefore, we agree with other studies proposing that the transmission and outbreaks of MRSA ST398 could be better studied using PFGE which showed a higher discriminatory power.15,16 All isolates contained the SCCmec type V, which is the most common SCCmec type identified in MRSA ST398.7
All isolates were resistant to tetracycline and carried the tetM and tetK genes. These genes, alone or in combination, are the most frequently identified in ST398 isolates.28 The resistance to tetracycline was highly expected and this resistance has been proposed as a marker to suspect the presence of MRSA ST398 in hospitals.
Regarding the macrolide-lincosamides resistance genes, ermC was the only erm gene detected. The unusual macrolide-lincosamide resistance phenotype EMS-CMR was identified in four MRSA in farm D being all classified as spa-type 1451, reflecting a local endemism of this resistance phenotype. The vga(A) gene was identified as the responsible for this resistance.29
Our work has several limitations. Neither swine were sampled when arriving at the four farms nor environmental samples were taken along the time when pigs were hosted in the farms. Therefore, we do not know the real influence of the cleaning procedures on the MRSA colonization of the swine. Besides, the resistance genes studied were limited we cannot exclude the presence of others. Moreover, only four farms were studied, our results may not be generalized to other swine farms.
Moreover, the adaptation of ST398 to the human being makes MRSA ST398 a matter of concern among humans and the public health representing a risk to themselves and the community.30 So, given the prevalence of swine MRSA colonization clinicians should be aware of the risk of MRSA colonization in people related with animal farming.
In summary, this study shows the MRSA colonization of pigs in indoor swine farms, being the MRSA prevalence lower in older swine. All MRSA were resistant to antimicrobials currently used in the clinical setting. The identification of a MRSA negative farm that used disinfecting and cleaning procedures suggests that appropriate hygienic practices could also be important in farming, as has been shown in the clinical setting.
Sources of financingThis work was financed by grants from Fondo de Investigaciones Sanitarias and Fondo Europeo para el Desarrollo Regional (FEDER) (07-0812) and Consejería de Sanidad de La Junta de Galicia (PS08/34).
Conflict of interestThe authors declare that there are not any conflicts of interest of any nature.
This work was partially presented at the 21st European Congress of Clinical Microbiology and Infectious Diseases (ECCMID) Milan, Italy.
We are grateful to farm workers and veterinarians for their help.