The etiological factors of colorectal cancer (CRC) are not precisely known, although genetic and environmental factors have been implicated. A possible association with Fusobacterium nucleatum may provide opportunities for an early diagnosis.
ObjectiveTo review studies that address the association between F. nucleatum and CRC.
MethodsThe MEDLINE PubMed database was searched using the terms «colorectal cancer» and “Fusobacterium nucleatum”, retrieving publications published up to January 1 2020. Stata software was used for a meta-analysis.
ResultsThe systematic review included 57 articles. Meta-analysis results indicated a more frequent presence of F. nucleatum in CRC tumour tissue samples in comparison to control samples of healthy tissue, with an odds ratio of 4.558 (95% CI: 3.312−6.272), and in comparison, to control samples of colorectal adenomas, with an odds ratio of 3.244 (95 % CI: 2.359−4.462).
ConclusionThere is a more frequent resence of F. nucleatum in the CRC. However, further studies are needed to verify this relationship.
Se desconocen los factores etiológicos exactos del cáncer colorrectal (CCR), aunque se ha intentado relacionar con factores genéticos y ambientales. La posible asociación con Fusobacterium nucleatum podría abrir posibilidades en el diagnóstico precoz.
ObjetivoRevisar los estudios que analizan la asociación entre F. nucleatum y el CCR.
MétodosSe utilizaron las publicaciones disponibles en la base de datos MEDLINE PubMed hasta el día 1 de enero de 2020, que incluían los términos «cáncer colorrectal» y «Fusobacterium nucleatum». Se realizó un metaanálisis con el software Stata.
ResultadosUn total de 57 artículos fueron incluidos en la revisión sistemática. El metaanálisis indicó una mayor presencia de F. nucleatum en muestras de tejido tumoral de CCR, con respecto a muestras control de tejido sano, con una odds ratio de 4,558 (IC 95%: 3,312–6,272), y cuando se utilizaron muestras control de adenomas colorrectales, con una odds ratio de 3,244 (IC 95%: 2,359–4,462).
ConclusiónHay una mayor presencia de F. nucleatum en el CCR. Sin embargo, se necesitan estudios que demuestren esta relación.
The aetiology of colorectal cancer (CRC) is multifactorial and includes genetic and epigenetic abnormalities1. Extrinsic factors such as intestinal dysbiosis might also play a role2; however, interindividual variability exists due to multiple genetic and environmental factors3. Hence, it is of special interest to investigate the possibility of a common microbial denominator.Fusobacterium nucleatum (FN) is among the most extensively studied bacteria4–6. It is a Gram-negative anaerobic bacterium that resides in the oral cavity as commensal microbiota, but it is also an opportunistic pathogen in periodontal diseases, primarily in gingivitis and periodontitis7. Its pathogenic power lies in its virulence factors, including: the presence of fimbriae, lipopolysaccharides, factors inhibiting chemotaxis of polymorphonuclear leukocytes and the production of toxic tissue metabolites8. However, its most significant virulence factor is FadA adhesin. This has been shown to be the greatest stimulant of inflammation as it creates a chronic proinflammatory environment that activates oncogenic signalling, thus stimulating epithelial cells9,10. Recent studies have demonstrated an increase in FadA in neoplastic CRC tissue, along with other proinflammatory markers such as COX-2, IL-8, IL-6, IL1ß and TNF-α11.
The possibility that FN is involved in the process of carcinogenesis could open up new avenues for early diagnosis. This justifies a comprehensive analysis of the current evidence on the relationship between FN and CRC. The objective of this study was to integrate the information available on the relationship between FN and CRC in a meta-analysis.
Material and methodsThe meta-analysis had a qualitative component (systematic review) and a quantitative component. The systematic review consisted of a description of the published studies, considering individual studies to be study “subjects”. A systematic search was conducted of all articles published in English or Spanish in journals indexed in MEDLINE, ISI Web of Knowledge and the Cochrane Library. The search terms used were “colorectal cancer” and “Fusobacterium nucleatum”. The selection criteria were the time limit (the search was restricted to all articles published in hard-copy and/or electronic format prior to January 2020), study type (research studies were selected) and language (the search was confined to articles written in English or Spanish). The following were excluded: studies on the relationship between FN and diseases other than CRC, studies on the relationship between CRC and bacteria other than FN, studies on the isolation of FN in species other than human beings, and reviews. The studies had to include patients with CRC, and they had to have the objective of directly or indirectly looking for FN and attempting to establish a possible association between FN infection and CRC. An internal quality system was used; this verified the reliability of the chosen method above, based on thorough review of each publication with tracing of the references in the publications to avoid missed studies.
The qualitative component referred to the statistical grouping of the results, with individual studies acting as research subjects, yielding, within and across publications, the 95% confidence interval, the odds ratio (OR) and the weights of the studies. The weighted mean was estimated by means of the random-effects model using DerSimonian and Laird’s method12, which is affected to a lesser extent by heterogeneity between studies. The inverse-variance test was used to assess heterogeneity. In addition, Higgins’s I2 inconsistency index was calculated; the value of this index indicates the percentage of variability due to heterogeneity between studies. Values over 75% indicate a strong heterogeneity level and would suggest a need to conduct further studies. Begg’s13 and Egger’s14 tests were used to detect publication bias. Data analysis was performed with the Stata 14 software programme.
ResultsSystematic reviewThe article search collected 199 publications; of them, 57 studies whose online version at least was published between 2011 and 2019 were selected (Table 1 and Fig. 1). Non-tumour area control samples were called Controls-1 (40 studies — 70.2%: 15 samples of adjacent non-tumour tissue, four faecal samples, two blood samples, one saliva sample, 18 non-tumour tissue samples). Adenoma samples were called Controls-2 (16%–28.1%). The Test-1 column indicates the test described by the authors to detect the micro-organism. In addition, some of the studies included the use of additional laboratory tests, called Test-2. Finally, the most salient conclusion of the research articles was also collected (“Article comments” column).
Systematic review of the literature published in the MEDLINE database and accessed via the PubMed interface until 1 January 2020 on the relationship between Fusobacterium nucleatum and colorectal cancer.
| Author | Year Physical publication | Total sex (f:m) | Cases CRC | Sex Cases CRC (f:m) | Control-1: No CRC (n) | Sex Control-1 | Description Group Control-1 | Control-2: Adenomas (n) | Sex Control-2 | What was analysed? | Test 1 | Test 2 | FN+/total Cases | FN+/total Control-1 | FN+/total Control-2 | Article comments |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Castellarin et al.15 | 2012 | NI | 11 | NI | 99 | NI | Adjacent non-tumour tissue | NI | NI | RNA | PCR | NI | 11/11 | 9/11 | NI | Higher rate of FN in CRC than in normal tissue |
| Rubinstein et al.16 | 2013 | NI | 51 | NI | 14 | NI | Tissue from tumour-free subjects | NI | NI | mRNA | PCR | NI | NI | NI | NI | Higher rate of FadA in CRC than in normal tissue |
| Kostic et al.4 | 2013 | NI | 27 | NI | 31 | NI | Tissue from tumour-free subjects | 28 | NI | DNA and RNA | PCR | FISH | 27/27 | 15/31 | 24/28 | FN creates a proinflammatory state that promotes progression to CRC |
| Warren et al.17 | 2013 | NI | 65 | NI | 65 | NI | Adjacent non-tumour tissue | NI | NI | rRNA | Metatranscriptomics | NI | NI | NI | NI | Higher rate of FN in CRC than in adjacent healthy tissue |
| Flanagan et al.18 | 2014 | NI | 122 | NI | 174 | NI | Adjacent non-tumour tissue | 52 | NI | DNA | PCR | NI | 70/122 | 33/174 | 12/52 | FN can be used as a marker for CRC |
| Mira-Pascual et al.19 | 2014 | NI | 7 | NI | 5 | NI | Tissue from tumour-free subjects | NI | NI | DNA and RNA | PCR | Pyrosequencing | 2/7 | 0/5 | NI | The increase in the levels of the bacteria studies was correlated with CRC |
| Tahara et al.20 | 2014 | NI | 149 | NI | 89 | NI | Adjacent non-tumour tissue | NI | NI | DNA | PCR | NI | 78/149 | 27/89 | NI | Higher rate of FN in CRC than in controls |
| Ito et al.21 | 2015 | NI | 511 | 225:286 | 20 | NI | Tissue from tumour-free subjects | 456 | 39:59 | DNA | PCR | NI | 286/511 | 3/20 | 142/456 | Higher rate of FN in CRC than in controls |
| Mima et al.22 | 2015 | NI | 598 | NI | 558 | NI | Adjacent non-tumour tissue | NI | NI | DNA | PCR | NI | 76/598 | 19/558 | NI | FN creates a proinflammatory state that promotes progression to CRC |
| Yu et al.10 | 2015 | NI | 42 | NI | 52 | NI | Adjacent non-tumour tissue | 47 | NI | DNA | PCR | NI | NI | NI | NI | Higher rate of FN in CRC than in controls |
| Fukugaiti et al.23 | 2015 | 4:13 | 7 | 2:5 | 10 | 2:8 | Faeces | NI | NI | DNA | PCR | FISH | 7/7 | 9/10 | NI | Higher rate of FN in CRC than in controls |
| Nosho et al.24 | 2016 | NI | 511 | 225:286 | NI | NI | No control tissue | NI | NI | DNA | PCR | NI | 44/511 | NI | NI | Lower rate of positive results for FN in the Japanese cohort |
| Repass et al.25 | 2016 | NI | 40 | NI | 40 | NI | Adjacent non-tumour tissue | NI | NI | DNA | PCR | NI | NI | NI | NI | Higher rate of FN in CRC than in normal tissue |
| Li et al.26 | 2016 | 46:55 | 101 | 46:55 | 101 | 46:55 | Adjacent non-tumour tissue | NI | NI | DNA | PCR | FISH | 88/101 | NI | NI | Larger amount of FN in CRC is associated with lower survival time |
| Yu et al.27 | 2016 | NI | 93 | NI | 20 | NI | Tissue from tumour-free subjects | 112 | NI | DNA | PCR | NI | 62/93 | 4/20 | 47/112 | FN might play a role in the serrated pathway of carcinogenesis for CRC |
| Mima et al.28 | 2016 | NI | 1,102 | NI | NI | NI | No control tissue | NI | NI | DNA | PCR | NI | 138/1,102 | NI | NI | Increases the rate of FN from CRC of the rectum to caecal carcinoma |
| Yu et al.5 | 2017 | NI | 74 | NI | 54 | NI | Tissue from tumour-free subjects | NI | NI | DNA | PCR | NI | 39/74 | 2/54 | NI | FN promotes CRC progression through modulation of autophagy pathways |
| Suehiro et al.29 | 2017 | NI | 158 | NI | 60 | NI | Tissue from tumour-free subjects | 19 | NI | DNA | PCR | NI | NI | NI | NI | Mean copies of FN: 17.5 in the control group, 122 in the adenoma group, 317 in the CRC group |
| Liang et al.30 | 2017 | NI | 203 | NI | 236 | NI | Tissue from tumour-free subjects | NI | NI | DNA | PCR | Metagenomic sequencing | NI | NI | NI | Higher rate of FN in CRC than in controls |
| Wong et al.31 | 2017 | NI | 104 | NI | 102 | NI | Tissue from tumour-free subjects | 103 | NI | DNA | PCR | NI | NI | NI | NI | Larger amount of FN in adenomas compared to controls p = 0.022 |
| Mehta et al.32 | 2017 | NI | 1,019 | NI | NI | NI | No control tissue | NI | NI | DNA | PCR | NI | 125/1,019 | NI | NI | The association between diet and CRC is modified by FN |
| Chen et al.33 | 2017 | NI | 98 | 40:58 | NI | NI | No control tissue | NI | NI | DNA | FISH | NI | 61/98 | NI | NI | Invasive FN activates the beta-catenin signalling pathway |
| Ye et al.34 | 2017 | NI | 25 | NI | 25 | NI | Adjacent non-tumour tissue | NI | NI | DNA | PCR | NI | 4/25 | 2/25 | NI | Higher rate of FN in CRC than in controls |
| Amitay et al.35 | 2017 | 220:280 | 46 | 15:31 | 231 | 121:110 | Tissue from tumour-free subjects | 223 | 84:139 | DNA | PCR | NI | 25/46 | 58/231 | 53/223 | Higher rate of FN in CRC than in controls |
| Park et al.36 | 2017 | 82:78 | 160 | 82:78 | NI | NI | No control tissue | NI | NI | DNA | PCR | NI | 107/160 | NI | NI | The larger amount of FN in tumour samples is correlated with the presence of intratumoural macrophages |
| Eklöf et al.37 | 2017 | NI | 39 | 19:20 | 65 | 30:35 | Tissue from tumour-free subjects | NI | NI | DNA | PCR | NI | 27/39 | 15/65 | NI | Larger amount of FN in CRC than in controls |
| Bullman et al.38 | 2017 | NI | 77 | NI | NI | NI | No control tissue | NI | NI | RNA | PCR | FISH | 45/77 | NI | NI | The reduction in FN with antibiotics causes less cancer proliferation |
| Drewes et al.39 | 2017 | NI | 58 | NI | 34 | NI | Tissue from tumour-free subjects | NI | NI | RNA | PCR | FISH | 17/58 | 1/34 | NI | Larger amount of bacteria in CRC than in healthy tissue |
| Yamaoka et al.40 | 2018 | NI | 100 | NI | 72 | NI | Adjacent non-tumour tissue | NI | NI | DNA | PCR | 75/100 | 46/72 | NI | Larger amount of FN in CRC than in controls | |
| Guevara et al.6 | 2018 | NI | 37 | NI | 37 | NI | Tissue from tumour-free subjects | NI | NI | Anti-FN antibodies | ELISA | NI | 37/37 | 12/37 | NI | The FN Fap2 protein is antigenic |
| Dai et al.41 | 2018 | 206:320 | 255 | 92:163 | 271 | 114:157 | Faeces | NI | NI | DNA | Metagenomics | NI | NI | NI | NI | Higher rate of FN in CRC than in controls |
| Liu et al.42 | 2018 | NI | 2,759 | NI | NI | NI | No control tissue | NI | NI | DNA | PCR | NI | 951/2,759 | NI | NI | Positive association between the empirical dietary inflammatory pattern (EDIP) and FN + CRC |
| Chen et al.43 | 2018 | NI | 25 | NI | 7 | NI | No control tissue | 8 | NI | DNA | FISH | Immunofluorescence | NI | NI | NI | Greater expression of TOX and CD4+ in FN − tissue versus FN + tissue. Negative correlation between FN abundance and TOX expression |
| Guo et al.44 | 2018 | NI | 215 | NI | 156 | NI | Tissue from tumour-free subjects | NI | NI | DNA | PCR | NI | NI | NI | NI | The FN/Bifidobacterium ratio had a sensitivity of 84.6% and a specificity of 92.3% in detecting CRC |
| Komiya et al.45 | 2018 | NI | 14 | NI | NI | NI | No control tissue | NI | NI | DNA | PCR | NI | 8/14 | NI | NI | FN samples in tumour tissue and saliva |
| Lee et al.46 | 2018 | NI | 246 | NI | NI | NI | No control tissue | NI | NI | DNA | PCR | NI | NI | NI | NI | High levels of FN are associated with lower survival in CRC metastases |
| Rezasoltani et al.47 | 2018 | NI | NI | NI | 31 | 15:16 | Tissue from tumour-free subjects | 87 | 33:54 | DNA | PCR | NI | NI | NI | NI | Significant relationship between polyp size and FN measurement |
| Hamada et al.48 | 2018 | 609:432 | 1,041 | 609:432 | NI | NI | No control tissue | NI | NI | DNA | PCR | NI | 135/1,041 | NI | NI | The association between FN and CRC varies depending on whether the CRC has an abnormal microsatellite pathway |
| Oh et al.49 | 2018 | 231:362 | 593 | 231:362 | NI | NI | No control tissue | NI | NI | DNA | PCR | NI | 204/593 | NI | NI | The tumour prognosis in relation to FN depends on the CRC location |
| Proença et al.50 | 2018 | NI | 43 | NI | 70 | NI | Adjacent non-tumour tissue | 27 | NI | DNA | PCR | NI | 33/43 | 23/70 | 14/27 | FN is a risk factor for CRC |
| Zhang et al.51 | 2019 | 41:53 | 94 | 41:53 | NI | NI | No control tissue | NI | NI | DNA | PCR | Microarray | 21/90 | NI | NI | FN induces expression of the BIRC3 gene in CRC |
| Saito et al.52 | 2019 | 30:51 | 24 | 7:17 | 10 | 7:3 | Tissue from tumour-free subjects | 47 | 16:31 | rRNA | PCR | NI | 14/24 | NI | 28/47 | FN may be associated with CRC and adenomas, and is a potential diagnostic marker for both |
| Guo et al.53 | 2019 | NI | 46 | NI | 36 | NI | Blood | NI | NI | DNA | Genomic sequencing | NI | NI | NI | NI | Greater presence of FN in tumour tissue than in blood samples |
| Feng et al.54 | 2019 | 13:12 | 15 | 8:7 | 10 | 5:5 | Tissue from tumour-free subjects | NI | NI | DNA | PCR | Western blot | 10/15 | 5/10 | NI | CREB protein expression is correlated with CRC metastasis whether in FN + or in FN− |
| Tunsjø et al.55 | 2019 | NI | 25 | NI | 22 | NI | Tissue from tumour-free subjects | 25 | NI | DNA | PCR | 16S rRNA sequencing | 15/25 | 5/25 | 4/22 | PCR for FN detection could be included as a CRC biomarker |
| Bundgaard-Nielsen et al.56 | 2019 | 141:132 | 99 | 55:44 | 76 | 35:41 | Adjacent non-tumour tissue | 96 | 51:47 | DNA | PCR | 16S rRNA sequencing | 23/99 | NI | 3/96 | There is no evidence that FN plays a role in the onset of adenomas, but it might play a role in the transition from adenoma to CRC |
| Guven et al.57 | 2019 | NI | 71 | NI | 77 | NI | Saliva from cancer-free controls | NI | NI | DNA | PCR | NI | NI | NI | NI | Larger amount of FN in saliva from patients with CRC compared to controls |
| Yachida et al.58 | 2019 | NI | 258 | NI | 291 | NI | Faeces | 67 | NI | DNA | Capillary electrophoresis | Metagenomic and metabolomic study | NI | NI | NI | FN abundance is correlated with tumour progression from intramucosal carcinoma to more advanced stages |
| Leung et al.59 | 2019 | 10:9 | 19 | 10:9 | 19 | 10:9 | Adjacent non-tumour tissue | NI | NI | DNA | PCR | NI | NI | NI | NI | Higher rate of FN in CRC than in normal tissue |
| Kunzmann et al.60 | 2019 | 60:130 | 190 | 60:130 | 190 | 60:130 | Adjacent non-tumour tissue | NI | NI | DNA | PCR | NI | 61/190 | 129/190 | NI | FN is more abundant in tumour tissue than surrounding tissue |
| Butt et al.61 | 2019 | NI | 485 | NI | 485 | NI | Blood | NI | NI | Anti-FN antibodies | Serology | NI | NI | NI | NI | Anti-FN antibodies have no association with the risk of developing CRC |
| De Carvalho et al.62 | 2019 | NI | 152 | NI | 57 | NI | Adjacent non-tumour tissue | NI | NI | DNA | PCR | NI | 35/152 | 6/57 | NI | Detection of FN DNA is associated with proximal tumour location; positivity for MSI or BRAF mutation; loss of MLH1, MSH2 or PMS2; and lower survival |
| Chen et al.63 | 2019 | 36:55 | 91 | 36:55 | NI | NI | No control tissue | NI | NI | DNA | PCR | NI | 25/91 | NI | NI | Larger amount of FN in CRC tissue is associated with lower survival time |
| Mima et al.64 | 2019 | 208:304 | 256 | 208:304 | NI | NI | No control tissue | NI | NI | DNA | PCR | NI | 140/256 | NI | NI | Larger amounts of FN and other bacteria associated with CRC increase the risk of anastomotic rupture following surgery |
| Liang et al.65 | 2019 | NI | 274 | NI | 353 | NI | Faeces | 353 | NI | DNA | PCR | Faecal immunohistochemistry test | 71/274 | 97/385 | 145/353 | Identification of m3 marker for CRC diagnosis |
| Haruki et al.66 | 2019 | 423:301 | 724 | 423:301 | NI | NI | No control tissue | NI | NI | DNA | PCR | NI | 99/724 | NI | NI | Inverse association between FN amount and tumour expression of BECN1 |
| Chen et al.67 | 2019 | NI | 148 | NI | NI | NI | No control tissue | NI | NI | DNA and RNA | PCR | FISH | 88/148 | NI | NI | FN is found in larger amounts in CRC with metastases |
CRC: colorectal cancer; FN: Fusobacterium nucleatum; NI: no information.
An increase in the number of studies published over the years was confirmed, but only in 2017 did this research significantly increase. The sex of the cases was reported in 20 (35.1%) studies overall, 11 (19.3%) Controls-1 studies and five (31.2%) Controls-2 studies. Until 2015, no studies reported the sex of the study participants. The most recent studies have tended to specify the sex of the study population. Analysis of the distribution of the studies with quantitative data found that such data were indicated in 40 (70.2%) Cases articles, 22 (38.6%) Controls-1 articles and 10 (25%) Controls-2 articles.
Quantitative analysisRegarding the number of positive results in the CRC samples, the highest rates of positive results were found in the studies by Castellarin et al.15, Kostic et al.4, Fukugaiti et al.23 and Guevara et al.6, in which all CRC samples analysed yielded a positive result in laboratory tests. By contrast, the lowest rate of positive results was found in a study by Nosho et al.24, in which just 8.6% of samples had a positive result. The number of samples for the Cases in each study varied widely. The lowest number was found in studies by Mira-Pascual et al.19 and Fukugaiti et al.23, with a total of seven cases analysed in each study. The largest sample size for the Cases corresponded to a study by Liu et al.42, with an analysis of 2759 samples. It should be noted that just four (7%) studies had more than 1000 individuals with CRC, and that they were conducted between 2017 and 2018.
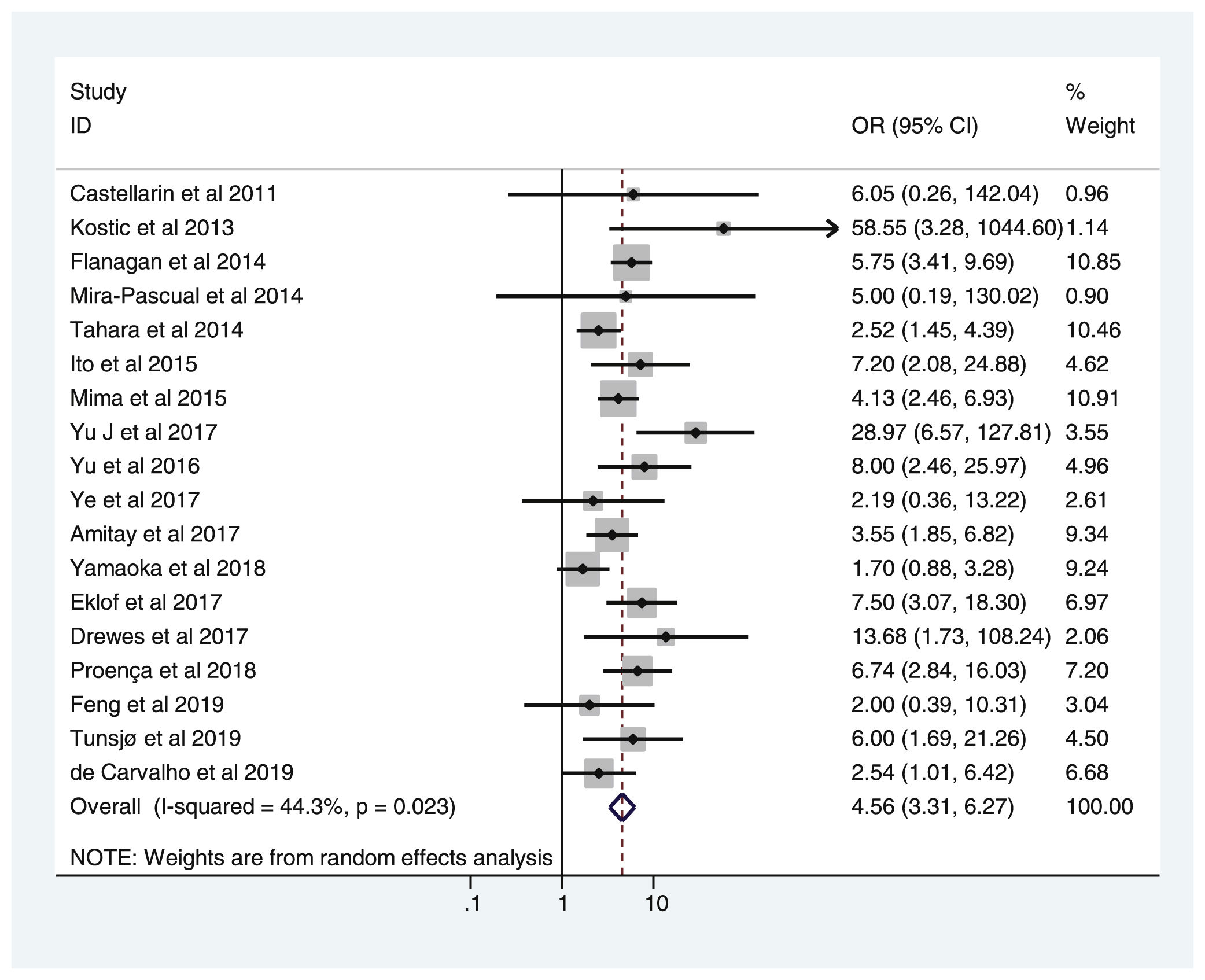
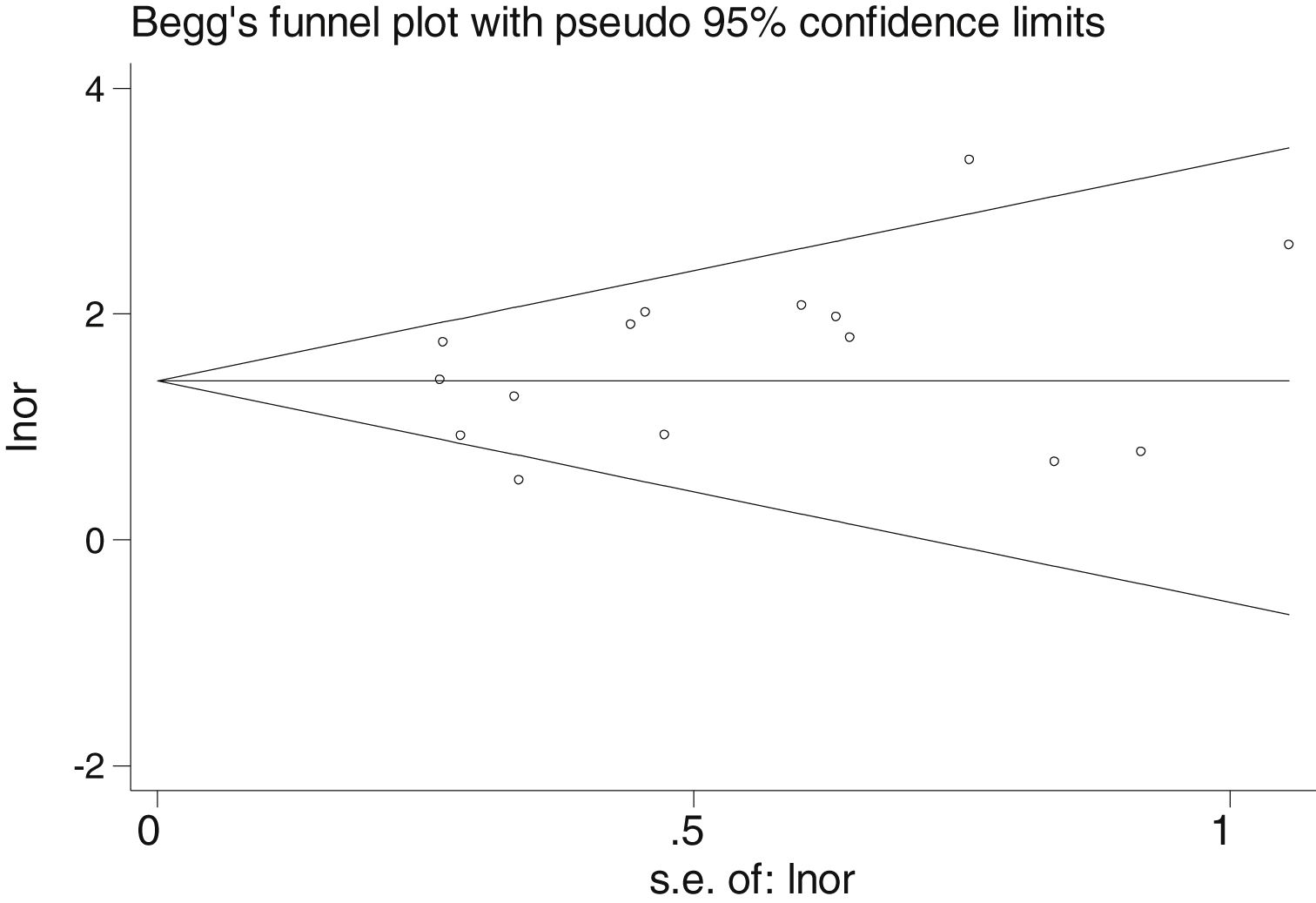
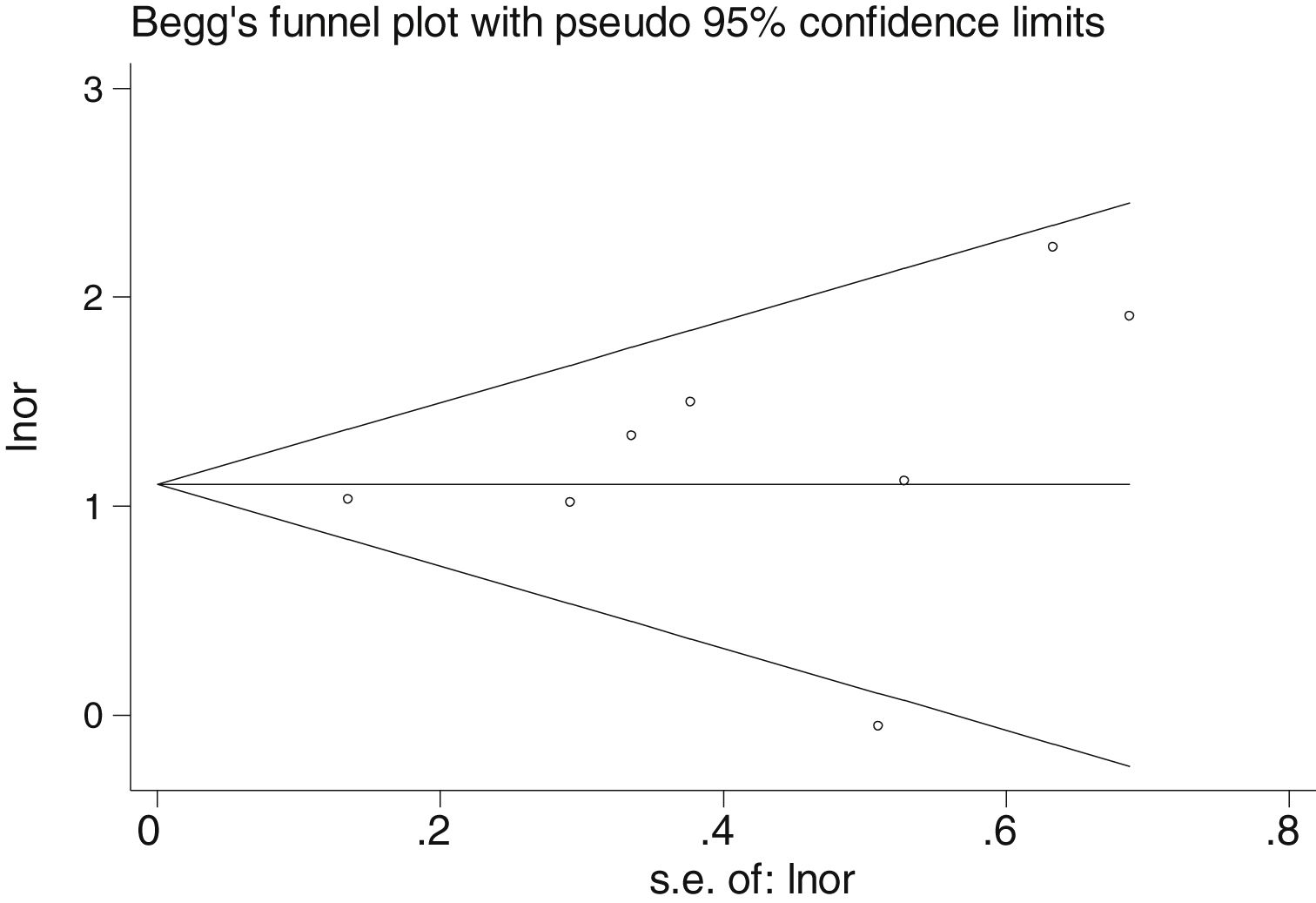
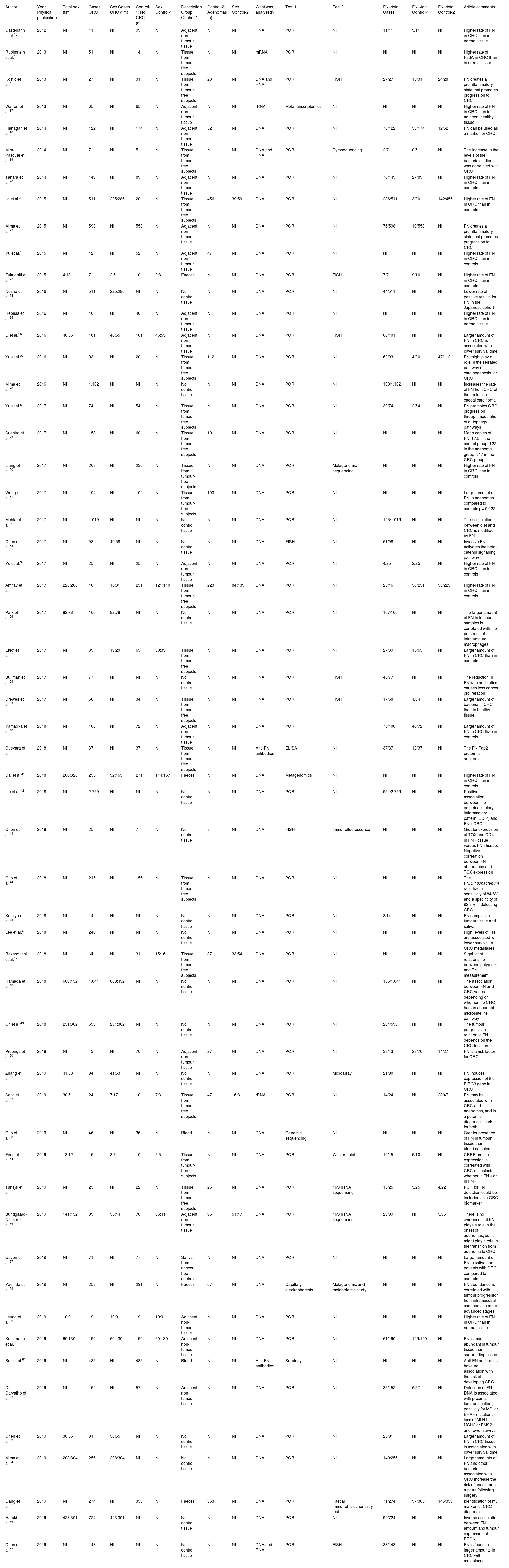
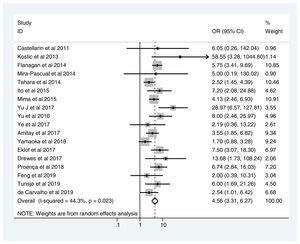
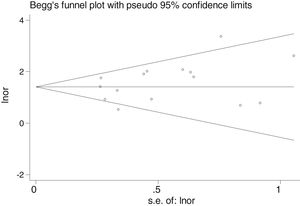
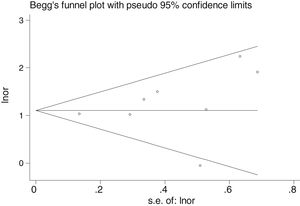
Table 2 shows the studies (18 out of 40, 45%) that compared, in Cases and Controls-1, positivity for FN infection markers with quantitative data from samples of colon tissue using molecular biology tests. The studies by Mima et al.28, Flanagan et al.18 and Tahara et al.20 were notable for their weight. All of them had a weight in excess of 10%. The overall OR estimate for the individual results obtained in these studies yielded a result of 4.558 (95% CI: 3.312−6.272), leading to the conclusion that there was a significant association (p < 0.001) between FN and CRC when Controls-1 were used. To calculate heterogeneity, the inverse-variance test was performed, with a value of χ2 exp. = 30.54, with 17 degrees of freedom and p = 0.023, meaning that the differences found between the studies were not due to chance. The heterogeneity study was corroborated using Higgins’s I2, with a result of I2=44,3% (Table 2 and Fig. 2). There was no obvious publication bias, as indicated in Begg’s and Egger’s tests, which were not significant (p = 0.767 and p = 0.210, respectively) (Fig. 3).
List of the 18 studies that compared, in Cases and Controls-1, positivity for Fusobacterium nucleatum infection markers in intestinal biopsy samples using molecular biology.
| Study | OR | 95% CI | % weight |
|---|---|---|---|
| Castellarin et al., 2011 | 6.053 | 0.258−142.040 | 0.96 |
| Kostic et al., 2013 | 58.548 | 3.282−1,044.602 | 1.14 |
| Flanagan et al., 2014 | 5.752 | 3.413−9.693 | 10.85 |
| Mira-Pascual et al., 2014 | 5.000 | 0.192−130.024 | 0.90 |
| Tahara et al., 2014 | 2.523 | 1.449−4.393 | 10.46 |
| Ito et al., 2015 | 7.203 | 2.085−24.884 | 4.62 |
| Mima et al., 2015 | 4.130 | 2.463−6.926 | 10.91 |
| Yu et al., 2017 | 28.971 | 6.567−127.806 | 3.55 |
| Yu et al., 2016 | 8.000 | 2.465−25.968 | 4.96 |
| Ye et al., 2017 | 2.190 | 0.363−13.219 | 2.61 |
| Amitay et al., 2017 | 3.551 | 1.850−6.815 | 9.34 |
| Yamaoka et al., 2018 | 1.696 | 0.876−3.282 | 9.24 |
| Eklöf et al., 2017 | 7.500 | 3.074−18.297 | 6.97 |
| Drewes et al., 2017 | 13.683 | 1.730−108.242 | 2.06 |
| Proença et al., 2018 | 6.743 | 2.838−16.026 | 7.20 |
| Feng et al., 2019 | 2.000 | 0.388−10.309 | 3.04 |
| Tunsjø et al., 2019 | 6.000 | 1.693−21.262 | 4.50 |
| De Carvalho et al., 2019 | 2.543 | 1.007−6.421 | 6.68 |
| D + L pooled OR | 4.558 | 3.312−6.272 | 100.00 |
Heterogeneity χ2 = 30.54 (gl = 17) p = 0.023.
I2 (variation in OR that can be attributed to heterogeneity) = 44.3 %.
Estimated variance between studies tau-squared = 0.1736.
Test of OR = 1: z = 9.31, p = 0.000.
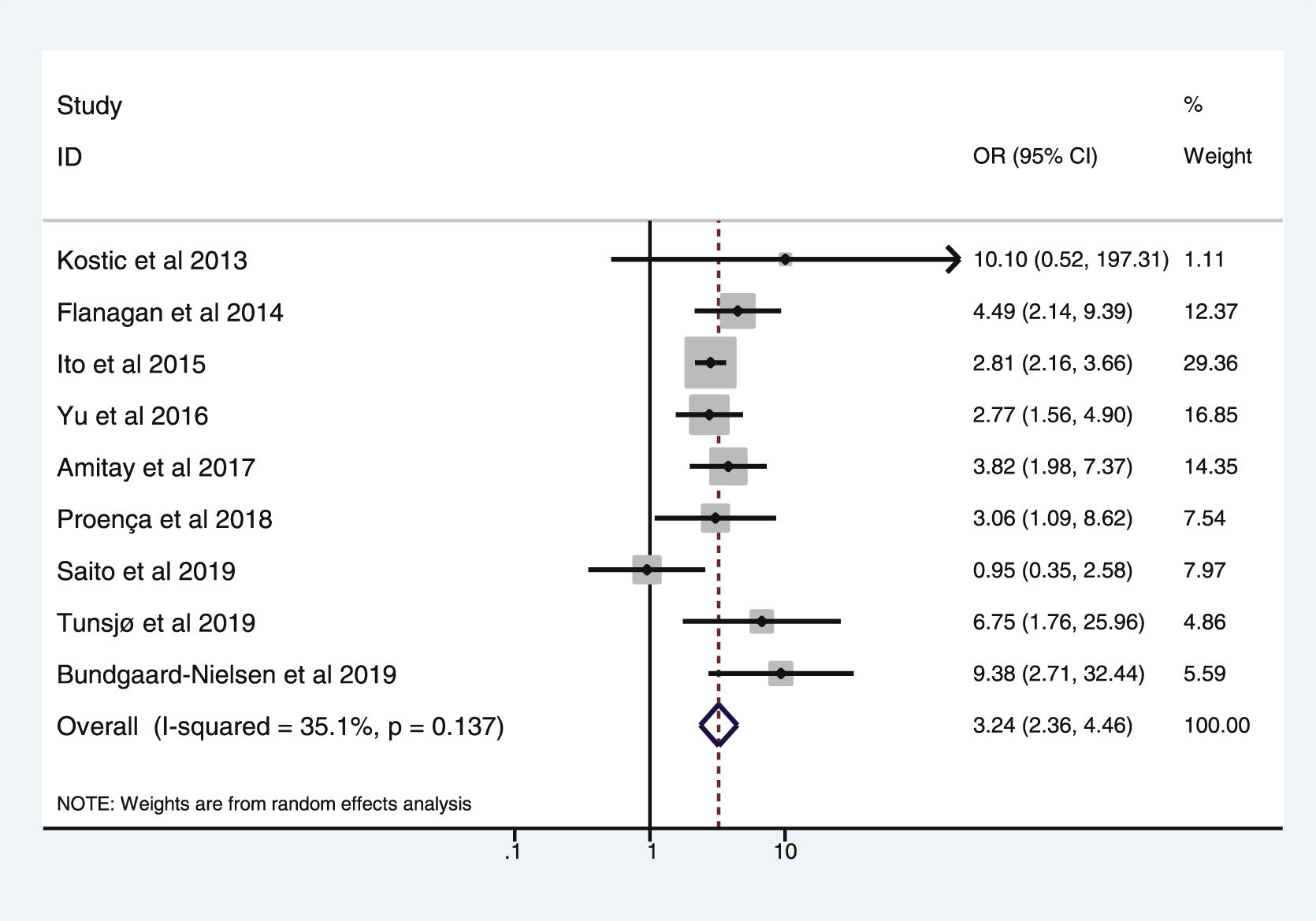
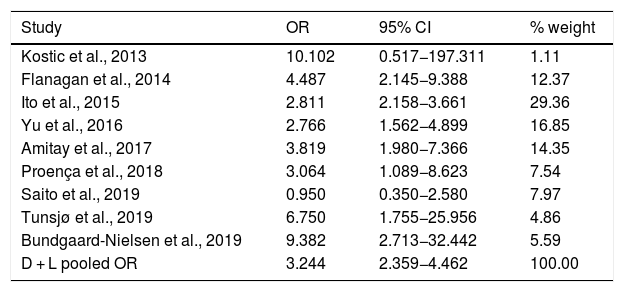
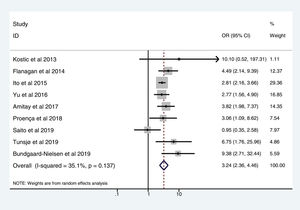
Table 3 shows the studies (nine out of 16, 56.25%) that compared, in Cases and Controls-2 (colorectal adenomas), positivity for genetic markers of FN infection with quantitative data from samples of colon tissue using molecular biology tests. The studies by Ito et al.21, Flanagan et al.18, Yu et al.27 and Amitay et al.35 all had notable weights, exceeding 12%. The overall OR estimate for the individual results obtained in these studies yielded a result of 3.244 (95% CI: 2.359−4.462), showing a statistically significant association between the presence of FN and CRC (p < 0.001). To analyse heterogeneity, the inverse-variance test was performed, with a value of χ2 exp. = 12.33, with eight degrees of freedom and p = 0.137, showing that there was homogeneity between the studies compared. In addition, Higgins’s I2 showed a value of 35.1%, indicating a low level of heterogeneity (Table 3 and Fig. 4).
List of nine studies that compared, in Cases and Controls-2, positivity for Fusobacterium nucleatum infection markers in intestinal biopsy samples using molecular biology.
| Study | OR | 95% CI | % weight |
|---|---|---|---|
| Kostic et al., 2013 | 10.102 | 0.517−197.311 | 1.11 |
| Flanagan et al., 2014 | 4.487 | 2.145−9.388 | 12.37 |
| Ito et al., 2015 | 2.811 | 2.158−3.661 | 29.36 |
| Yu et al., 2016 | 2.766 | 1.562−4.899 | 16.85 |
| Amitay et al., 2017 | 3.819 | 1.980−7.366 | 14.35 |
| Proença et al., 2018 | 3.064 | 1.089−8.623 | 7.54 |
| Saito et al., 2019 | 0.950 | 0.350−2.580 | 7.97 |
| Tunsjø et al., 2019 | 6.750 | 1.755−25.956 | 4.86 |
| Bundgaard-Nielsen et al., 2019 | 9.382 | 2.713−32.442 | 5.59 |
| D + L pooled OR | 3.244 | 2.359−4.462 | 100.00 |
Heterogeneity χ2 = 12.33 (gl = 8) p = 0.137.
I2 (variation in OR that can be attributed to heterogeneity) = 35.1%.
Estimated variance between studies tau-squared = 0.0719.
Test of OR = 1: z = 7.24 p = 0.000.
There was no obvious publication bias, as indicated by Begg's and Egger's tests, which were not significant (p = 0.108 and p = 0.441, respectively) (Fig. 5).
DiscussionThis study found that the relationship between FN and CRC is a topic of growing interest, given the exponential rise in studies that have explored this relationship. With regard to the sample size of the studies, 31 (54.4%) had fewer than 100 case samples, and just four (7%) had more than 1000 samples. Similarly, in the studies published, just 22 (55%) had numerical data for Controls-1 and 10 (17.5%) had numerical data for Controls-2. Efforts have been made in recent years to correct this, however, and the most current studies have exhibited a trend towards publishing the corresponding values. Above all, future studies must report quantitative data for FN positivity in samples, as this constitutes solid evidence subject to statistical processing on the association thereof with CRC. Also important is the influence of sex on the results, which has scarcely been studied in this regard, given that such data have been omitted from many studies. The breakdown of this relationship by sex could shed light on whether sex could be considered a risk factor in relation to the presence of FN, since non-transmissible chronic diseases are related to lifestyle factors in men and women. In this systematic review, the sex of the patients was only indicated in 20 (35.1%) studies out of the total of 57 articles collected.
Similarly, future studies could examine other variables, such as diet. Some studies, including the study by Hussan et al.68, have collected data for this variable and found a fibre-rich diet to be linked to a lower risk of developing CRC despite having a larger number of FN-positive samples in the colon mucosa, which would seem contradictory.
It should also be noted that the majority of the samples analysed in the study were tissue samples. That is to say, the CRC samples were tumour tissue samples and the control samples were samples of healthy tissue, since, in theory, the study thereof should offer more complete information. From what has been published to date, it has been deduced that FN has a significant presence in CRC tissue compared to adjacent healthy tissue15,18,20,28,69, with a gradual increase in FN from healthy mucosa to adenoma and from adenoma to adenocarcinoma. This suggests that FN may play an important role as of the earliest stages of colon cancer onset4,10,18,70, that there is an association between the presence of FN and lymph node invasion by CRC15,26,71 and that FN accompanies tumour cells in metastases38. Thus it has been explained that the amount of FN in CRC is linked to stage, chemotherapy resistance, higher recurrence and lower survival, with said amount acting as an independent predictive factor10,26,28,38,70,72,73.
Just four (7%) studies tested faecal samples, one (1.8%) tested blood samples and one (1.8%) tested saliva samples, concluding that there is an overabundance of FN in the faeces of patients with CRC. FN is 132 times more abundant in the faeces of patients with CRC, and its determination has been identified as a useful marker for detecting CRC29–31, showing a sensitivity of 80.2% and a specificity of 80.7%, with these figures increasing to 92 %–93% when it is combined with faecal occult blood detection74. It would be interesting to confirm the association between FN and CRC simultaneously in samples of tissue, faeces, saliva and blood. Should research be intensified and a significant association discovered, it would have a great deal of diagnostic utility for patients, since these samples are relatively easy to collect with no need for a colonoscopy to acquire them.
Based on all studies conducted to date, the involvement of FN in the mechanism of carcinogenesis in CRC could be agreed upon, since there is a statistically significant association between the presence of FN and CRC. However, in order to determine that this bacterium is the true origin of CRC, the causality criteria, primarily temporality, direction and association, must be met75. In this case, the temporality criterion, by which the effect must be preceded by the cause, could not be demonstrated. Most studies that have investigated the association between FN and CRC have been retrospective, meaning it could not be known whether the bacterium was present before the tumour developed or was acquired later; more evidence, then, is needed to confirm this causality.
Multiple studies, including those by Mima et al.28, Flanagan et al.18 and Tahara et al.20, which are notable for their weight in the meta-analysis, confirmed that there is a relationship between FN and CRC in comparison of healthy tissue from Controls-1 and CRC Cases. However, this association must be clarified for reasons including the fact that, at present, no prospective studies have found the presence of the bacterium to precede the onset of cancer. It is now known that the aetiopathogenic substrate corresponds to the activation of a chronic inflammatory state resulting from adhesion of the bacterium through a mechanism of intestinal dysbiosis. This entire process leads to an initiation of pathways associated with colorectal carcinogenesis, notably the Wnt/beta-catenin pathway, whose dysregulation causes failures in cell growth and tumour progression76. In-depth examination of the activation of these proinflammatory pathways and the mechanisms by which they occur will open up multiple possibilities for diagnosis and treatment in one of the highest incidence cancers today, with substantial benefits in the course of this disease. Our meta-analysis found FN to be more abundant in colorectal cancer tissue samples compared to healthy tissue with an OR of 4.558 and a 95% confidence interval of 3.312−6.272, confirming the significant association between the presence of FN in the tissue samples with the development of CRC. However, the meaning of this relationship must be clarified with prospective studies that confirm the temporality of the association. Other meta-analysis such as Hussan et al.’s68 have also pointed to this need.
In addition, recent studies such as Yu et al.’s70 have described the role of FN in chemoresistance in patients with CRC, indicating that an increased amount of FN in tumour tissue is linked to a higher rate of chemoresistance. For this reason, it would be useful to conduct further studies on survival time in patients in relation to FN levels in tumour tissue, since this could signify the existence of more therapeutic targets in this disease such as bacteria-eradicating treatment.
In the case of the comparison between the Controls-2 (colorectal adenomas) and the CRC Cases, it is important to note that not the all articles used samples of the same types of colorectal adenomas; a wide variety of histological types falling under the label of “colorectal adenoma” were studied. Furthermore, at present, fewer studies have linked the presence of FN to colorectal adenomas. The scarcity of articles, coupled with the lack of uniform criteria for sample selection, indicates that this association, as well as the question of whether FN contributes less to an adenoma forming and more to it becoming malignant, should be studied in greater depth in the future.
Multiple articles, including but not limited to Mima et al.28, Yu et al.5 and Lee et al.46, have found a relationship between CRC survival and the presence or absence of FN, though no studies have found FN eradication to lead to a better prognosis in cancer treatment or an improvement in chemotherapy. In addition, no studies have linked good dental hygiene to CRC. FN is a type of bacteria that colonises the oral cavity, and therefore the relationship between FN amounts in the oral cavity and in the colon mucosa could be examined.
The main limitation of this systematic review was that it included studies published in MEDLINE, accessed via PubMed, and not in other databases or doctoral dissertations on the topic. Hence we assume that published information, however limited, might have been missed; on the other hand, such information could be presumed to be of little significance. Regarding the language limitation, only studies published in English or Spanish were included, although most of the journals indexed in MEDLINE were published in English, and only two studies were excluded for this reason. Even the association of higher rates of FN in CRC samples does not imply a causality that could point to a preventive strategy. It could stem from facilitation by tumour tissue of subsequent FN infection.
ConclusionsBased on the results of this systematic review and meta-analysis, we found that there is an association between the presence of FN and CRC. Lastly, to arrive at a definitive conclusion, further comparative studies must be conducted in sufficient numbers of patients using a combination of multiple microbiological techniques for individual subjects and samples, with simultaneous analysis of neoplastic tissue and healthy tissue by means of standardised techniques with suitable sensitivity.
Conflicts of interestThe authors declare that they have no conflicts of interest.
Please cite this article as: Villar-Ortega P, Expósito-Ruiz M, Gutiérrez-Soto M, Ruiz-Cabello Jiménez M, Navarro-Marí JM, Gutiérrez-Fernández J. La asociación entre Fusobacterium nucleatum y el cáncer colorrectal: una revisión sistemática y metaanálisis. Enferm Infecc Microbiol Clin. 2022;40:224–234.