Severe acute infectious diarrhoea is a serious public health problem worldwide. In diarrhoea of bacterial origin, except for that caused by Clostridioides difficile, most are invasive in nature1 and antibiotic therapy may be necessary in some specific situations. Knowing the susceptibility to different classic and alternative antimicrobials is therefore crucial, as the number of episodes of enteritis caused by multidrug-resistant pathogens is increasing.2–5 The aim of this study was to analyse the rate of sensitivity to antibiotics, as well as the ability to predict the sensitivity of Salmonella to ciprofloxacin through susceptibility to nalidixic acid.
We conducted a retrospective descriptive study on the results of the stool cultures recorded in our Laboratory Information System due to episodes of enteritis from January 2016 to December 2019. The geographical area was the province of Granada and the population in scope was the Hospital Universitario Virgen de las Nieves catchment area, with a basic reference population of 330,486, but which provides tertiary healthcare. The population treated came from specialised care (including hospital in-patients, outpatient clinics and the Accident and Emergency department). Express information on symptoms was lacking. Non-solid faeces were transported and processed following a strict working protocol.1 The MicroScan system (Beckman Coulter, Barcelona, Spain) was used to determine the sensitivity of rapidly growing microorganisms. The minimum inhibitory concentration (MIC) of azithromycin and fosfomycin was determined by diffusion gradient (MIC Test Strip, Liofilchem®, Italy). When there was a breakpoint, results were interpreted following the guidelines of the European Committee on Antimicrobial Susceptibility Testing (2020) (https://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/v_10.0_Breakpoint_Tables.pdf). Absolute and relative frequencies were calculated for the categorical variables, and measures of central tendency and dispersion for the numerical variables. We used the χ2 test for the statistical analysis of categorical variables and Student's t-test to analyse the numerical variables, such as age. The data were analysed with the IBM® SPSS® Statistics program.
For this study, there were 9290 negative stool cultures and 1160 microorganisms were isolated from 1080 (10.4%) different episodes. In general, Campylobacter (additional Table 1 in the Appendix A) was the most isolated (P < .001), and Campylobacter jejuni was the most common. The year with the most positive stool cultures was 2017 (P < .001), followed by 2019 (additional Table 2 in the Appendix A). The rate of Campylobacter increased in the last year (P = .008); the Salmonella rate was highest in 2017 (P < .001) and Aeromonas remained stable (P = .52). Campylobacter and Salmonella were more common in summer and autumn (P = .002 and P < .001, respectively). There were more isolates in the child population than in adults (60.2% vs 39.8%), despite there being more adult samples (69.3% vs 30.7%), so the yield of stool culture is higher in the child population (15% vs 4%; P < .001). Overall, 64.8% of Campylobacter (P < .001), 54% of Salmonella (P = .009) and 61.1% of Aeromonas were detected in the child population (P = .002). In patients under a year old, Campylobacter was the most isolated species, followed by Aeromonas. In the adult population, the most commonly isolated pathogens were the same as in the paediatric population (additional Table 3 in the Appendix A). In most of the patients, a single pathogen was detected, in 56 patients two were isolated simultaneously; of these, in 96.4% of the episodes, one of the microorganisms was Aeromonas. The most common accompanying pathogen in co-infection with Aeromonas was Campylobacter. Of the co-infections with Aeromonas, 85.2% occurred in children, primarily under one year of age.
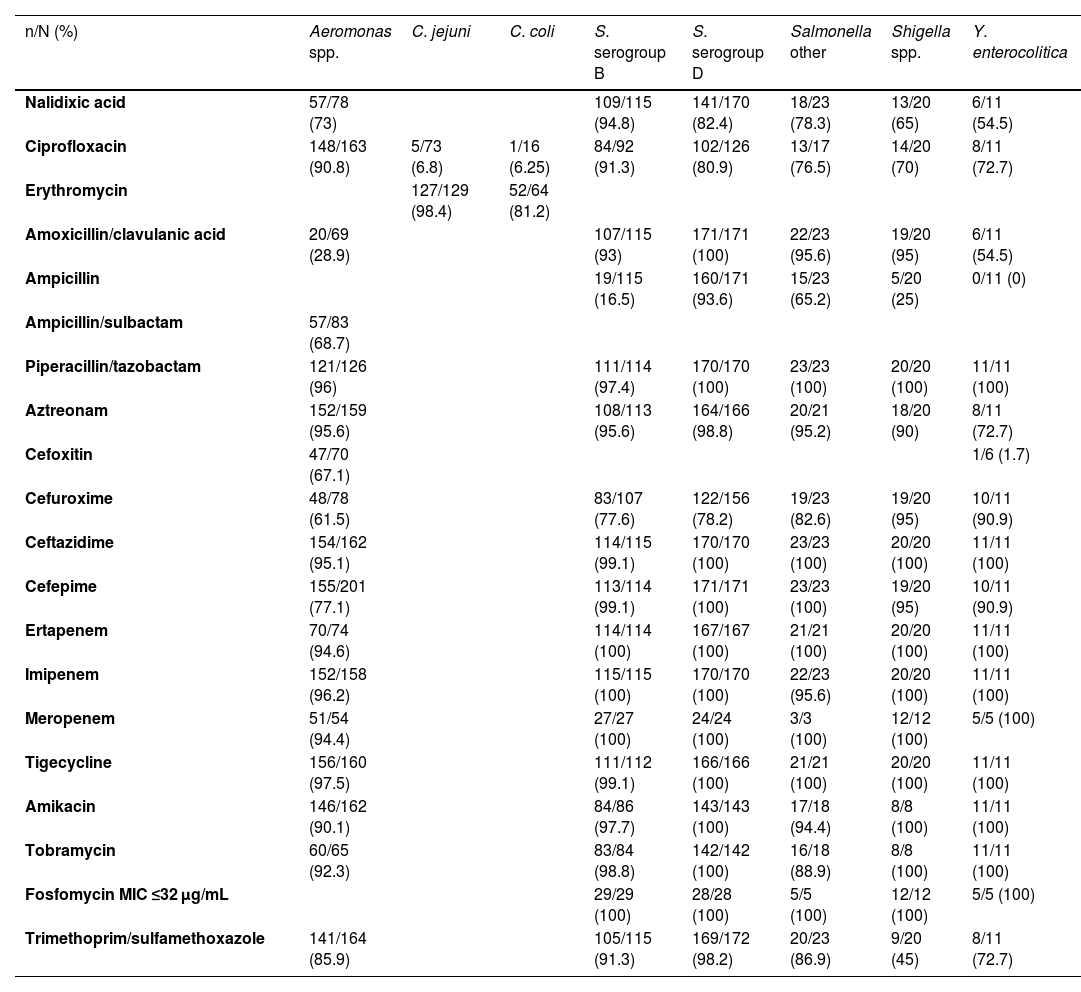
The antibiotic sensitivity is shown in Table 1. Interesting is the low sensitivity of Campylobacter to ciprofloxacin, given that in some cases of severe acute diarrhoea empirical treatment with quinolones is started, and the fact that Campylobacter jejuni is the most detected microorganism in stool cultures. The results of the sensitivities to nalidixic acid and ciprofloxacin in Salmonella are shown in additional Tables 4 and 5 in the Appendix A. In general, susceptibility to nalidixic acid quite accurately predicted susceptibility to ciprofloxacin, except in Salmonella enterica serogroup B, possibly due to the presence of non-classic quinolone resistance phenotypes in S. enterica serogroup B. For the study of susceptibility to azithromycin, 133 isolates from stool cultures were taken from 2016 to 2019 (additional Tables 6-8 in the Appendix A). In the fosfomycin susceptibility analysis, we recovered 31 Campylobacter coli isolates, noting that the MICs were high (additional Table 9 in the Appendix A).
Antibiotic sensitivity of the isolates.
| n/N (%) | Aeromonas spp. | C. jejuni | C. coli | S. serogroup B | S. serogroup D | Salmonella other | Shigella spp. | Y. enterocolitica |
|---|---|---|---|---|---|---|---|---|
| Nalidixic acid | 57/78 (73) | 109/115 (94.8) | 141/170 (82.4) | 18/23 (78.3) | 13/20 (65) | 6/11 (54.5) | ||
| Ciprofloxacin | 148/163 (90.8) | 5/73 (6.8) | 1/16 (6.25) | 84/92 (91.3) | 102/126 (80.9) | 13/17 (76.5) | 14/20 (70) | 8/11 (72.7) |
| Erythromycin | 127/129 (98.4) | 52/64 (81.2) | ||||||
| Amoxicillin/clavulanic acid | 20/69 (28.9) | 107/115 (93) | 171/171 (100) | 22/23 (95.6) | 19/20 (95) | 6/11 (54.5) | ||
| Ampicillin | 19/115 (16.5) | 160/171 (93.6) | 15/23 (65.2) | 5/20 (25) | 0/11 (0) | |||
| Ampicillin/sulbactam | 57/83 (68.7) | |||||||
| Piperacillin/tazobactam | 121/126 (96) | 111/114 (97.4) | 170/170 (100) | 23/23 (100) | 20/20 (100) | 11/11 (100) | ||
| Aztreonam | 152/159 (95.6) | 108/113 (95.6) | 164/166 (98.8) | 20/21 (95.2) | 18/20 (90) | 8/11 (72.7) | ||
| Cefoxitin | 47/70 (67.1) | 1/6 (1.7) | ||||||
| Cefuroxime | 48/78 (61.5) | 83/107 (77.6) | 122/156 (78.2) | 19/23 (82.6) | 19/20 (95) | 10/11 (90.9) | ||
| Ceftazidime | 154/162 (95.1) | 114/115 (99.1) | 170/170 (100) | 23/23 (100) | 20/20 (100) | 11/11 (100) | ||
| Cefepime | 155/201 (77.1) | 113/114 (99.1) | 171/171 (100) | 23/23 (100) | 19/20 (95) | 10/11 (90.9) | ||
| Ertapenem | 70/74 (94.6) | 114/114 (100) | 167/167 (100) | 21/21 (100) | 20/20 (100) | 11/11 (100) | ||
| Imipenem | 152/158 (96.2) | 115/115 (100) | 170/170 (100) | 22/23 (95.6) | 20/20 (100) | 11/11 (100) | ||
| Meropenem | 51/54 (94.4) | 27/27 (100) | 24/24 (100) | 3/3 (100) | 12/12 (100) | 5/5 (100) | ||
| Tigecycline | 156/160 (97.5) | 111/112 (99.1) | 166/166 (100) | 21/21 (100) | 20/20 (100) | 11/11 (100) | ||
| Amikacin | 146/162 (90.1) | 84/86 (97.7) | 143/143 (100) | 17/18 (94.4) | 8/8 (100) | 11/11 (100) | ||
| Tobramycin | 60/65 (92.3) | 83/84 (98.8) | 142/142 (100) | 16/18 (88.9) | 8/8 (100) | 11/11 (100) | ||
| Fosfomycin MIC ≤32 μg/mL | 29/29 (100) | 28/28 (100) | 5/5 (100) | 12/12 (100) | 5/5 (100) | |||
| Trimethoprim/sulfamethoxazole | 141/164 (85.9) | 105/115 (91.3) | 169/172 (98.2) | 20/23 (86.9) | 9/20 (45) | 8/11 (72.7) |
The percentage of antibiotic sensitivity has been calculated based on the sensitive isolates versus the total isolates of each bacteria tested for the antibiotic.
In conclusion, Campylobacter jejuni is the most commonly isolated microorganism, followed by Salmonella, which is most notable in the child population. Interestingly, erythromycin was the first choice antibiotic against Campylobacter. Also striking was the high degree of resistance of Salmonella serogroup B to ampicillin. Nalidixic acid sensitivity predicted the susceptibility of Salmonella to ciprofloxacin, except in the case of S. enterica serogroup B. Azithromycin can be considered a good option for empirical treatment of choice, or an alternative in the case of multidrug resistance to other antibiotics, in severe acute enteritis caused by the main enteroinvasive bacteria, apart from Shigella.
Funding- •
Open access financial support: Universidad de Granada/Consorcio de Bibliotecas Universitarias de Andalucía (CBUA) [University of Granada/Consortium of Andalusian University Libraries].