Accumulating evidence has demonstrated that long non-coding RNAs (lncRNAs) play important regulatory roles in the tumorigenesis and progression of gastric cancer (GC). The aim of this study was to construct the prognostic predictive model of lncRNAs signature and improve the survival prediction of GC.
Patients and methodsThe expression profiling of lncRNAs in large GC cohorts was performed from The Cancer Genome Atlas (TCGA) databases using the lncRNAs-mining approach, including training data set (N=160) and testing data set (N=159). A 13-lncRNAs signature significantly associated with overall survival (OS) in the training data set was selected. The prognostic value of this 13-lncRNAs signature was then confirmed in the test validation set and the entire validation set, respectively.
ResultsBased on lncRNA expression profiling of 319 patients with stomach adenocarcinoma (STAD), prognostic 13-lncRNAs signature was found to be significantly associated with the prognosis of GC. Compared to patients with low-risk scores, patients with high-risk scores had a significantly shorter survival time. Moreover, functional enrichment analysis indicated that this 13-lncRNAs signature was potentially involved in multiple biological processes, such as DNA replication and cell cycle signaling pathway.
ConclusionsThe prognostic model of the 13-lncRNAs signature established by our study could improve the survival prediction of GC to a greater extent.
Las pruebas acumuladas demostraron que los ARN no codificantes de larga duración (ARNlC) desempeñaban los importantes papeles reguladores en la tumorigénesis y la progresión del cáncer gástrico (CG). El objetivo de este estudio fue construir el modelo predictivo de pronóstico de la firma de los lncRNA y mejorar la predicción de supervivencia del GC.
Pacientes y métodosEl perfil de expresión de los lncARN en grandes cohortes de GC se realizó a partir de las bases de datos del Atlas del Genoma del Cáncer (TCGA) utilizando el enfoque de minería de lncARN, incluyendo el conjunto de datos de entrenamiento (N=160) y el conjunto de datos de pruebas (N=159). Se eligió la firma de 13 lncARN significativamente asociada con la supervivencia general (OS) en la serie de capacitación. El valor pronóstico de esta firma de 13-lncARN se confirmó luego en la serie de validación de pruebas y en toda la serie de validación, respectivamente.
ResultadosBasado en el perfil de expresión de lncRNA de 319 pacientes con adenocarcinoma de estómago (STAD), se encontró que la firma de 13-lncRNA de pronóstico estaba significativamente asociada con el pronóstico de GC. En comparación con los pacientes con puntuaciones de bajo riesgo, los pacientes con puntuaciones de alto riesgo tuvieron un tiempo de supervivencia significativamente más corto. Además, el análisis de enriquecimiento funcional indicó que esta firma de 13-lncARN estaba potencialmente involucrada en múltiples procesos biológicos, como la replicación del ADN y la vía de señalización del ciclo celular.
ConclusionesEl modelo de pronóstico de la firma de 13-lncARN establecido por nuestro estudio podría mejorar mejor la predicción de supervivencia del GC.
Gastric cancer (GC) is the most commonly diagnosed cancer in the Eastern Asia and the third leading cause of cancer morality worldwide.1 Considerable efforts have been make to improve the therapeutic treatment of GC. Unfortunately, clinical outcome and long-term survival are still not satisfactory, which is mainly attributed to the failure of early detection and survival prediction of GC. Patients with GC are commonly presented with non-specific symptoms and diagnosed at the advanced stage in daily clinical practice. Some people are reluctant to undergo gastroscopy, considering that it is an invasive examination. The current serological prognostic biomarkers, such as carcinoembryonic antigen (CEA) and carbohydrate antigen (CA) 19-9, have a low specificity and sensitivity for prediction, especially in early stage of GC.2 Thus, there is an urgent need to explore the reliable and appropriate molecular biomarkers for the prediction of prognosis in patients with GC.
Up to date, particular attention has been paid to the newly discovered long non-coding RNAs (lncRNAs), which are defined as transcripts longer than 200 nucleotides without apparent potential of protein coding.3 Interestingly, lncRNAs play the important regulatory roles to modify the expression of protein-coding genes at transcriptional, post-transcriptional and epigenetic levels.4 The reported lncRNAs with aberrant expression were increasing exponentially in GC, in correlation with the processes of proliferation, immortality, angiogenesis, motility and viability.5–8 It had been proved that certain lncRNAs could be served as potential diagnostic, prognostic and therapeutic biomarkers in GC. Nevertheless, the systematic understanding of lncRNAs in GC is still not well illustrated, particularly for combined lncRNAs signature on long-term survival of patients with stomach adenocarcinoma (STAD).
A large population-based analysis in the screening of prognosis-associated lncRNAs was performed using the whole-transcriptome RNA-sequencing (RNA-seq) data from The Cancer Genome Atlas (TCGA) database. The purpose of this study was to construct the prognostic predictive model of combined lncRNAs signature served as STAD-specific biomarkers. Findings in this study may help to provide a comprehensive bioinformatics picture with important implications of personalized therapies in GC, eventually bringing clinical benefits to the patients.
Materials and methodsTCGA lncRNA dataset and patient informationLncRNA expression profiles and clinical data on STAD cohort were available at TCGA data portal (https://portal.gdc.cancer.gov/projects). Patients were chosen for prognostic model building if they met the following criteria: (1) lncRNA expression profile, complete clinicopathological and follow-up data all available; (2) OS of more than 30 days. After filtering, a total of 319 patients were enrolled for further analysis. In addition, these lncRNAs appeared in >70% of the total samples with an average count >1. The patients were further randomly assigned to the train set (n=160, used to identify key lncRNAs) and the test set (n=159, used to verify the lncRNA signature). Altogether, the lncRNA profiles were acquired for all the patients, which were standard normalized within and among the samples. The final expression level of each lncRNA was defined as the log2(x+1) of the raw expression level.
Prognostic model construction and statistical analysisUnivariable Cox regression analysis was performed using the computing environment R with Survival package. Identified target lncRNAs were considered statistically significant if their p values were less than or equal to 0.05. We further made a selection of target lncRNAs to pick out the prognosis-related lncRNA signature. Robust likelihood-based survival analysis was performed by using R with Rbsurv package. Each identified prognosis-related lncRNA gene was fitted in the univariable Cox regression model of the train set and obtained the corresponding parameter. The log-likelihood of each lncRNA corresponding parameter was further evaluated in the validation set. A series of predictive models were constructed according to the above described procedure. The prognosis-related lncRNAs were strictly selected based on this model. After fitting all candidate predictive models in to Akaike information criterions (AICs), the optimal predictive model was selected by the lowest AIC value.
Risk score formula establishment and validationThe risk score formula was established by these prognosis-related lncRNAs and weighted by their estimated regression coefficients in the multivariable Cox regression analysis of the train set. Based on this formula, the risk score for each patient in the train set was calculated. Meanwhile, patients were classified into high-risk score or low-risk score groups by using the corresponding median risk score as the cut-off point. The receiver operating characteristic (ROC) curve was obtained to predict OS at 3 and 5 years by using R with survival ROC package. The optimal cut-off point was chosen by the maximal combination of sensitivity and specificity. Survival difference between the low-risk and high risk group were assessed by the Kaplan Meier curve and compared by the log-rank test of the multivariable analyses. The risk score formula was further validated by fitting into the validation set and the entire set. The prognostic predictive performance was measured by the AUC values from time-dependent ROC analysis. Moreover, the accuracy of the risk score was assessed to predict OS at 3 and 5 years. The multivariable Cox proportional hazards regression was performed to evaluate the independence of these thirteen STAD-specific lncRNAs. Stratification analysis of common clinical characteristics, such as tumor stage and patient gender, were conducted. All statistical analyses were conducted using R language (Version 3.3.3). Survival curves and ROC curves were generated by the “survival” (version 2.41-3), and “survivalROC” (version 1.0.3) packages.
Bioinformatic analysis of lncRNA target genes and pathwaysThe correlation networks between these thirteen prognosis-related lncRNAs and potential target genes were investigated by Spearman's test. Moreover, gene ontology (GO) and KEGG pathway enrichment was used to analyze the biological process of target genes and pathways by Cluster Profiler. The enriched function annotations of GO terms and KEGG pathways were considered significantly when the p value is less than 0.05.
ResultsSTAD patients in the train and test setA total of 319 patients from the TCGA database were included in this study, which were randomly divided into training set (N=160) and testing set (N=159). Finally, 14,448 lncRNAs were identified from TCGA STAD database. Among these acquired lncRNAs, 5674 lncRNAs have a mean FPKM of >0.1. Baseline demographic and clinical characteristics of the two groups did not differ statistical significantly (p>0.05). These characteristics are summarized in Table 1.
Characteristics of the study population in the train set and the validation set.
| Characteristics | Train set(n=159) | Test set(n=160) | Entire set(n=319) |
|---|---|---|---|
| Age (mean±SD) | 65.18±10.44 | 65.34±10.58 | 65.26±10.51 |
| Sex (male/female) | 104/56 | 100/59 | 204/115 |
| Clinical stage (I/II/III/IV) | 21/53/69/17 | 24/52/67/16 | 45/105/136/33 |
| Status (deceased/alive) | 61/99 | 71/88 | 132/187 |
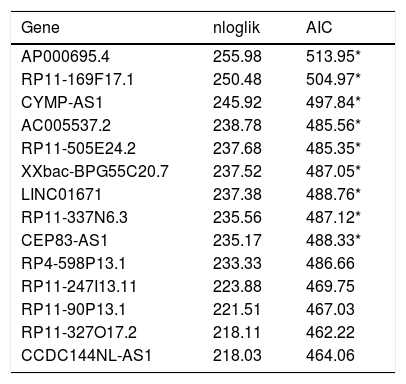
To define the association of lncRNAs with prognosis of STAD patients, we used univariable Cox regression analysis to identify 365 lncRNAs, which were significantly associated with overall survival (OS) of STAD patients (p<0.05). Robust likelihood-based survival analysis was used to picked out prognostic 13-lncRNAs signature from these 365 lncRNAs (Table 2).
The prognostic models signature in the training set (N=160).
| Gene | nloglik | AIC |
|---|---|---|
| AP000695.4 | 255.98 | 513.95* |
| RP11-169F17.1 | 250.48 | 504.97* |
| CYMP-AS1 | 245.92 | 497.84* |
| AC005537.2 | 238.78 | 485.56* |
| RP11-505E24.2 | 237.68 | 485.35* |
| XXbac-BPG55C20.7 | 237.52 | 487.05* |
| LINC01671 | 237.38 | 488.76* |
| RP11-337N6.3 | 235.56 | 487.12* |
| CEP83-AS1 | 235.17 | 488.33* |
| RP4-598P13.1 | 233.33 | 486.66 |
| RP11-247I13.11 | 223.88 | 469.75 |
| RP11-90P13.1 | 221.51 | 467.03 |
| RP11-327O17.2 | 218.11 | 462.22 |
| CCDC144NL-AS1 | 218.03 | 464.06 |
nloglik is the abbreviation of negative log-likelihoods, AIC is the abbreviation of Akaike information criterion.
To facilitate the application of these lncRNAs in clinical practice, we designed a risk prediction formula and calculated the risk score for each patient in the train set. The 160 STAD patients in training set were classified to the high-risk group (N=80) and low-risk group (N=80) by the median risk score. The optimal cut-off point for the classification of differential survival outcome for STAD was identified when risk score is 1.26. Obviously, patients in the low-risk group showed the better outcome than those in the high-risk group by Kaplan–Meier analysis (p<0.001, Fig. 1A).
Construction of the thirteen-lncRNAs signature in the training set. (A) Kaplan–Meier overall survival (OS) analysis in the training set between patients in the high-risk group and low-risk group. (B) The distribution of risk score, OS and lncRNA expression for the patients in the training set. Columns represent patients.
The distribution of the risk scores, OS, survival status, and corresponding lncRNA expression profiles of the 160 patients in the train set were shown in Fig. 1B. These thirteen STAD-specific lncRNAs tended to be more highly expressed in the high-risk group. Notably, the high-risk group comprised 4 patients with death and 34 patients without death, whereas the low-risk group comprised 15 patients with death and 65 patients without death. However, the difference between two groups in OS was marginally significant (chi-square test, p=0.003).
Validation of thirteen-lncRNAs signature in the test set and entire TCGA setUsing the same risk score formula and threshold value in train set, patients in test set (N=159) and entire TCGA set (N=319) were classified into high-risk groups and low-risk groups. With convinced evidence that patients in high-risk group of testing set (n=79) had poor outcome than those in the low-risk group by Kaplan–Meier analysis (p=0.003, Fig. 2A). Similar results were still observed in entire TCGA set (p<0.0001, Fig. 2B).
Validation of the thirteen-lncRNAs signature in the testing set and entire set. (A) Kaplan–Meier overall survival (OS) analysis in the testing set between patients in the high-risk group and low-risk group. (B) Kaplan–Meier overall survival (OS) analysis in the entire set between patients in the high-risk group and low-risk group. (C) The distribution of risk score, OS and lncRNA expression for the patients in the testing set. (D) The distribution of risk score, OS and lncRNA expression for the patients in the entire set. Columns represent patients.
The distributions of the risk scores, OS, survival status, and corresponding lncRNA expression profiles in the test set and entire set of patients were shown in Fig. 2C and D (ranked according to increasing risk scores). Likewise, these thirteen risky lncRNAs were downregulated in the low-risk group and upregulated in the high-risk group. Moreover, the majority of patients with death in test set and entire TCGA set were clustered in the high-risk group. In the test set, high-risk group comprised 46 patients with death and 34 patients without death, whereas the low-risk group comprised 25 with death and 55 without death. The difference in OS between two groups was statistically significant (chi-square test, p<0.001). Similarly, the high-risk group comprised 85 patients with death and 74 patients without death in the entire TCGA set, while the low-risk group comprised 47 patients with death and 113 patients without death. Consistent with testing set, the difference in OS between two groups of entire set was significant (chi-square test, p<0.001).
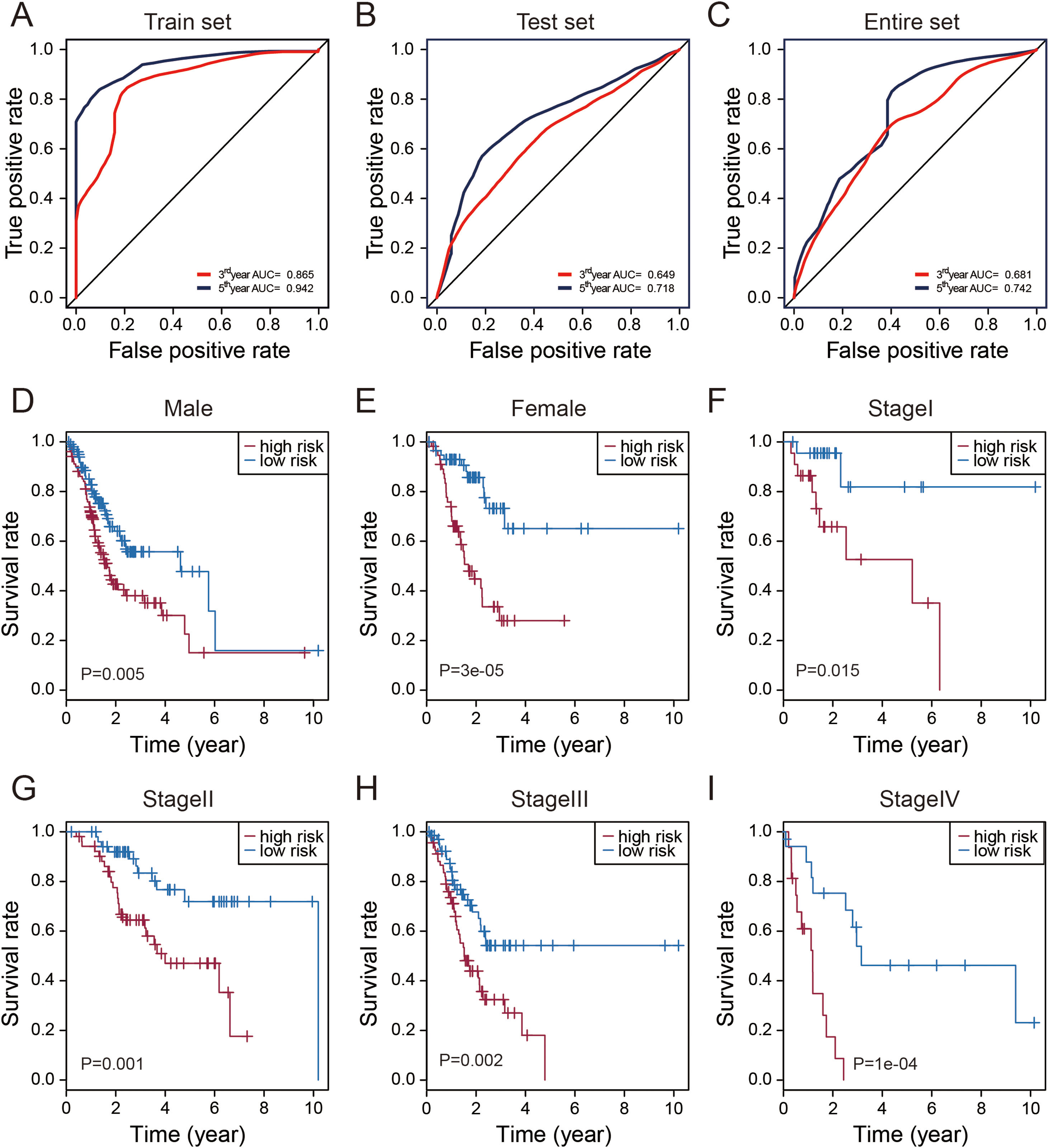
Evaluation of the predictive performance of the thirteen-lncRNAs signatureTime-dependent receive operating characteristic (ROC) curve analysis was performed to evaluate the prognostic performance of these thirteen STAD-specific lncRNAs. It was demonstrated that these thirteen prognosis-related lncRNAs achieved area under the curve (AUC) values of 0.865 and 0.942 for predicting prognosis in the train set at 3 and 5 years (Fig. 3A). The AUC values were 0.649 and 0.718 in the test set at 3 and 5 years (Fig. 3B) and 0.681 and 0.742 in entire set at 3 and 5 years (Fig. 3C). All of the AUC values exceed 0.6, indicating that these thirteen prognosis-related lncRNAs were performed well for the prediction of prognosis in STAD patients.
Performance assessment and comparison of the thirteen-lncRNAs signature by survival ROC and stratification analyses. (A–C) The Receiver operating characteristic (ROC) analysis of overall survival (OS) for the thirteen-lncRNAs signature in training set, testing set and entire set. (D) Kaplan–Meier curves for male patients. (E) Kaplan–Meier curves for female patients. (F) Kaplan–Meier curves for patients with stage I. (G) Kaplan–Meier curves for patients with stage II. (H) Kaplan–Meier curves for patients with stage III. (I) Kaplan–Meier curves for patients with stage IV.
Stratification analysis of tumor stage and patient gender was conducted to determine whether these thirteen STAD-specific lncRNAs maintain their prognostic value in the different context of common clinical features. All 319 patients were stratified by tumor stage into a stage I (N=45), stage II (N=105), stage III (N=136) and IV dataset (N=33). Using this 13-lncRNAs signature, patients in the stage I dataset were classified into a high-risk group (N=23) or a low-risk group (N=22); these groups had significantly different OS (p=0.015, Fig. 4F). Likewise, the patients in the stage II, III and IV datasets also were classified into a high-risk group and a low-risk group, which also differed significantly in OS (p<0.005, Fig. 4G–I). Similarly, all 319 patients were stratified by gender into the male dataset (N=204) and female dataset (N=115). Patients in the male dataset could be stratified into high-risk group (N=102) and low-risk group (N=102) with significant difference in OS (p=0.005, Fig. 4C). The analogous result was obtained in the female dataset (p=0.00003; Fig. 4D).
Functional enrichment analysis of prognostic lncRNAs1476 potential target genes of these thirteen lncRNAs were first identified by Go terms and KEGG pathways functional enrichment analysis. The top 10 enriched KEGG pathways were shown in Fig. 4A. It was demonstrated that a series of cancer-related pathways were highly activated in STAD patients, such as DNA replication and cell cycle signaling pathway. The enriched GO terms mainly included ncRNA processing and ribosome biogenesis, which had long been recognized as functions of lncRNAs. The top 10 enriched GO terms of biological process were shown in Fig. 4B, suggesting that these thirteen prognostic lncRNAs were tightly correlated with the carcinogenesis of gene expression and critical cell biological functions.
DiscussionPrognostic thirteen-lncRNAs signature was established for survival prediction of STAD by the comprehensive analysis of TCGA database. This lncRNAs signature was reproducible and reliable in another large-scale and independent cohort of STAD, supporting its value and effectiveness. Our present study explored the potential impact of combined lncRNAs signature to predict the prognosis of GC. Another notable finding was the lncRNAs-mining approach described here which could potentially be applied and served as a useful method for the systematic identification of combined lncRNAs signature in the clinical practice. Moreover, functional enrichment analysis of lncRNAs was further performed to elucidate the target genes and pathways.
Long non-coding RNA (lncRNA) is one kind of non-coding RNA which is longer than 200 nt and lack of protein-coding ability.9 LncRNAs were considered as “trash RNA” without arousing much attention in the past. Recently, accumulating evidence demonstrated that lncRNAs were involved in the multiple of cancer biological processes, such as tumor initiation, growth, metastasis and multidrug resistance.10 For instance, lncRNA CCAL expression was upregulated in GC tissues and the CCAL/miR-149/FOXM1 axis exerted functions as a key regulator in the metastasis of GC.11 Elevated UFC1 promoted GC progression by regulating miR-498/Lin28b signaling pathway.12 HOXC-AS3 was significantly upregulated in GC tissues compared with the corresponding non-tumor tissues, which would be served as an independent predictor for the overall survival in GC. In addition, HOXC-AS3 regulated cell proliferation and migration both in vitro and in vivo. RNA-seq analysis for whole transcriptome studies indicated HOXC-AS3 played an important role in the tumorigenesis of GC. The activated function of HOXC-AS3 was mediated by the interaction with YBX1.13 Recent studies had identified one kind of new lncRNA GMAN, regulating the translation of EFNA1 mRNA by binding competitively to GMAN-AS RNA, which was increased in GC tissues and associated with tumor metastases.14
With the development of microarray and high-throughput technology, thousands of tumor-specific prognostic lncRNAs were found and selected. The emerging role of lncRNAs were involved in gastric cancer and acted as regulators at the transcriptional or post-transcriptional level. Some lncRNAs predict negative prognosis and exhibits oncogenic activity in gastric cancer. Some lncRNAs had been found with up-regulated expression in gastric cancer, such as ABHD11-AS1,15 GAPLINC,16 Ak058003,16 ANRIL,17 H19,18 whereas down-regulated expression of lncRNAs in gastric cancer, like GACAT1,19 GAS5,20 LINC00982.21 However, most previous studies of lncRNA were designed by a general point of view, whereas the integrated alteration pattern of lncRNA was ignored, resulting in no assurance of finding more putative biomarkers. Similar to miRNAs signature of GC, a combined lncRNAs signature may substantially improve the prediction of clinical outcome.22,23 In our present study, 13 differentially expressed lncRNAs were all associated with poor prognosis. Better understanding of the roles of lncRNAs in GC may provide new biomarkers for early diagnosis and prognostic evaluation. Therefore, further study of this 13-lncRNAs signature could provide more information about putative biomarkers in the screening of GC.
In the previous study by Ren W, five lncRNAs (CTD-2616J11.14, RP1-90G24.10, RP11-150O12.3, RP11-1149O2 3.2, and MLK7-AS1) were identified in 76 gastric cancer-specific lncRNAs, which explored the potential of combining lncRNAs signature to predict the prognosis of GC.24 The risk score of those five lncRNAs was an independent predictor of overall survival in patients with gastric cancer. However, our ideal prognostic predictors model of GC was constructed by the combination of 13-lncRNAs signature, using the risk score formula in our study. Interestingly, the reason why there were no overlapping lncRNAs of our 13-lncRNAs signature with the above mentioned 5-lncRNAs signature, which could be explained by the molecular heterogeneity and methodology used. In the comprehensive analysis of lncRNA-sequencing data, this 13-lncRNAs signature could effectively divide patients into high-risk and low-risk groups with significantly different OS. We then successfully validated the relation of this 13-lncRNAs signature with prognosis of gastric cancer patients in the testing set and the entire set, indicating good reproducibility and reliability for the prediction of prognosis. Moreover, stratification analysis of tumor stage and patient gender was conducted to find out the prognostic value of 13-lncRNAs signature in the different context of common clinical features.
Interestingly, there were two RP11 lncRNAs in their 5 prognostic-related lncRNAs,24 and six RP11 lncRNAs in our 13 prognostic-related lncRNAs. To our knowledge, the functions of this 13-lncRNAs signature have not been reported. Majority of these novel lncRNAs do not even have the official name. Several RP11 lncRNAs have been reported in previous gastric cancer studies, such as RP11-1149O23.2. It had been proved that the mechanism of RP11-1149O23.2 in the cis-regulation of TNFRSF10A, suggesting a potential role of RP11-1149O23.2 in the process of programmed cell death.24 Similarly, RP11-357H14.17 might be involved in the progression of diffuse-type gastric cancer by altering cell migration and invasion.25 RP11-119F7.4 was more frequently downregulated in the gastric cancer.26 Moreover, RP11-363E7.4 was mainly involved in the PI3K signaling pathway and in the regulation of the actin cytoskeleton, indicating lncRNA RP11-363E7.4 may affect gastric cancer through above these two pathways.27 In addition, several other RP11 lncRNAs have been considered as a potential prognostic biomarker in other kind of cancer studies, like RP11-650L12.2 in colorectal cancer,28 RP11-445H22.4 in breast cancer29 and RP11-766N7.4 in esophageal squamous cell carcinoma.30 According to the current research, the RP11 lncRNAs mentioned above were correlated with tumorigenesis, invasion depth, metastasis, tumor size and stage. Importantly, these 6 RP11 lncRNAs were identified by this study, which may have the same potential and should be verified by experiments in further research.
The initiation and progression of GC is a long-term process, involving the activation of key signaling pathways and dysregulation of cellular processes. Functional enrichment analysis of this prognostic 13-lncRNAs signature showed that most enriched biological processes and pathways were implicated in the development of GC. In our study, the enriched GO terms were mainly included extracellular matrix organization and extracellular structure organization. While commonly known for degradation of the extracellular matrix, matrix metalloproteinases (MMPs) exhibit broad potential in the treatment of cancers. LncRNAs HOTAIR and HOXC-AS3 were reported to play the important roles in stabilizing or degrading the extracellular matrix, especially in correlation with GC metastasis. Above these lncRNAs could regulate extracellular matrix degradation by modulating the expression of metastasis-associated cancer genes, such as ICAM-1, MMP1, MMP2, MMP3 and MMP9.
ConclusionsWe would like to acknowledge some certain limitations of this study. Undoubtedly, the clinical information of some patients was incomplete, which may influence the assessment of the predictive model and reduce the robustness of this study. In addition, independent cohorts from multicenter study in a large population are required to validate the prognostic value of this 13-lncRNAs signature before it can be applied to clinical practice. Despite some limitations, this combination of 13 STAD-specific lncRNAs signature could improve survival prediction and guide the tailored therapy for patients with GC.
Authors’ contributionH.W., T.C.Z., Z.Z., and H.L. conceived and designed this study. T.C.Z. and Z.Z. collected and assembled data. Z.M.Y., F.X.T., X.J.Z. and H.K.T. analyzed and interpreted data. H.W., H.L. and Z.Z. drafted the manuscript. Z.Z., H.L. and T.C.Z. prepared figures and tables. All authors read and confirmed the final version of this manuscript.
FundingThis work was supported by the National Natural Science Foundation of China (Grant Number: 81860433 and 81960359), the Natural Science Youth Foundation of Jiangxi Province (Grant Number: 20192BAB215036), the Foundation for Fostering Young Scholar of Nanchang University (Grant Number: PY201822).
Conflict of interestNone.