Notch receptors regulate many aspects of metazoan development and tissue renewal, such as binary cell-fate decisions, survival, proliferation and differentiation, in different cell types and in a context dependentmanner. In hematopoiesis, Notch signalling actively contributes to Tcell development by driving hematopoietic stem cell- (HSC-) derived progenitors seeding the thymus into the T-cell lineage, while simultaneously avoiding alternative cell fates. According to this critical role, deregulated Notch signalling has important consequences in T-cell generation, survival and proliferation during thymopoiesis, and significantly contributes to the generation of T-cell acute lymphoblastic leukemias (T-ALL). Therefore, understanding Notch-dependent molecular pathways that control physiological and pathological development of T-cell progenitors within the thymus has become an intensive area of research in recent years. Several genes and signalling cascades, including c-myc, NF-κB and PI3K, have been identified as critical targets involved in Notch-induced T-cell oncogenesis, and others such as interleukin-7 receptor (IL-7R) have recently been suggested to participate in the process. In this review, we highlight recent studies on Notch that reveal new molecular details about how Notch signalling guides human thymic immigrants along the T-cell lineage and how deregulated activation of Notch can contribute to T cell leukemogenesis, in part by directly regulating expression of the IL-7R.
Los receptores Notch regulan diferentes aspectos del desarrollo de los metazoos y de la renovación tisular, que incluyen decisiones binarias de destino celular, supervivencia, proliferación o diferenciación, en diferentes tipos celulares y tejidos y en diferentes contextos celulares. En hematopoyesis, la señalización por Notch contribuye activamente al desarrollo de los linfocitos T, induciendo un programa madurativo específico en los progenitores que llegan al timo procedentes de las células madre hematopoyéticas (HSC) de la médula ósea y bloqueando, simultáneamente, la generación de linajes celulares alternativos. Conforme a esta importante función, la desregulación de la señalización por Notch tiene consecuencias críticas para la supervivencia y proliferación de las células T y contribuye significativamente a la generación de la leucemia T linfoblástica aguda (TALL). Por tanto, el estudio de las vías moleculares inducidas por Notch que determinan la maduración de los progenitores T, así como su transformación oncogénica durante el desarrollo intratímico, constituye una importante área de investigación en la actualidad. Se ha demostrado la participación de varios genes y rutas de señalización, tales como c-myc, NF-κB y PI3K, en la oncogénesis de las células T inducida por Notch y más recientemente se ha sugerido la implicación del receptor de la interleucina 7 (IL-7R) en el proceso. En esta revisión discutiremos recientes estudios moleculares que revelan cómo Notch determina la generación de linfocitos T a partir de los inmigrantes intratímicos y cómo su desregulación puede contribuir a la generación de leucemias T-ALL, en parte debido a la regulación directa de la expresión del IL-7R.
Hematopoietic stem cells (HSCs) give rise to all blood lineages, except T-cells, in a limited number of specialized niches within the fetal liver or the adult bone marrow. T lymphocytes are a unique exception, as their development takes place in a dedicated lymphoid organ, the thymus. The generation of T lymphocytes from HSCs-derived precursors that seed the thymus is an orchestrated process specifically controlled by the thymic microenvironment(1-5). Within the thymus, both interactions with surface molecules expressed by thymic epithelial cells (TECs) and response to soluble factors produced by TECs are mandatory events, which enable the delivery of signals that are essential for directing early thymic progenitors (ETPs) along the T-cell linage. Specifically, interactions of Notch receptors with their ligands expressed on TECs(6, 7) and signalling mediated through the interleukin 7 receptor (IL-7R) in response to IL-7 produced by TECs(8, 9) are crucial events that regulate thymopoiesis in both mouse and man.
Notch signalling is required both to promote T-cell specification and commitment of multipotent progenitors (MPPs) that seed the thymus and to block intrathymic development of alternative cell fates, including myeloid, B or natural killer (NK) lineages(1, 2, 10-14) (Figure 1). Once Tcell specification is achieved, survival and proliferation of T-cell precursors is induced at two successive checkpoints. First, by signalling through the IL-7R, which enables expansion of the intrathymic pool of T-cell precursors(8,9 15–19), and later on by the TCRβ-pTα pre-T cell receptor (pre-TCR)(20, 21), which promotes the selection and proliferation of pre-T cells with a successful rearrangement at the TCRβ locus (Figure 2). Interestingly, several studies have pointed toward a prominent role of Notch signalling in the regulation of the latter proliferation events(21, 22), while others have suggested the implication of Notch in the former(14). Moreover, Aster and co-workers(23) recently provided evidence that Notch-mediated signalling is crucial for the development of T-cell acute lymphoblastic leukemia (T-ALL), as they found that more than 50% cases of human T-ALLs have activating NOTCH1 mutations. Therefore, understanding the molecular basis that control Notch-dependent function in T-cell precursors and their malignant counterparts is an important issue that offers the opportunity of developing novel targeted therapies against this type of cancer. Aimed at improving our knowledge of Notch l function on normal and leukemogenic human Tcell development, we have recently approached gain- and loss-of-function studies that have provided evidence of a crucial interplay between Notch l and IL-7R pathways(24) , which underscores the molecular bases of Notch1-IL-7R functional interactions in physiology and pathology and represents the focus of this review.
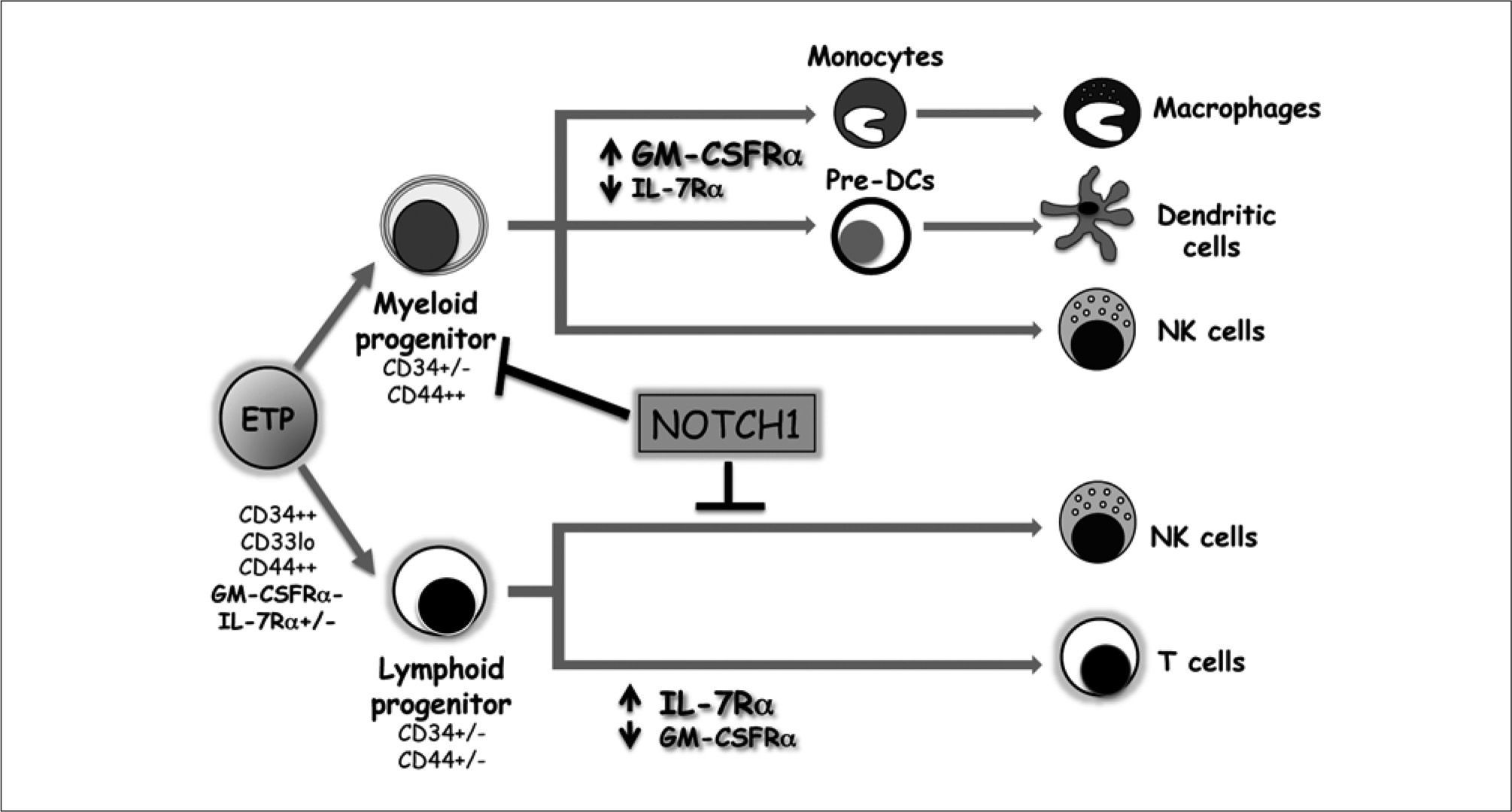
Proposed model of lymphoid and myeloid differentiation pathways in the human thymus. Early thymic progenitors (ETPs) seeding the thymus from bone marrow are multipotent, as they can differentiate into both myeloid- (dendritic cells, macrophages and natural killer (NK) cells) or lymphoidlineage cells (T cells and NK cells), through intermediate myeloid- or lymphoid-primed progenitors, respectively. The latter corresponds to the conventional T/NK bipotent progenitor. Notch1 signaling promotes T-cell differentiation from ETPs by blocking the generation of intermediate myeloid progenitors thus favoring the lymphoid cell fate. Thereafter, Notch1 signalling blocks NK development from bipotent T/NK progenitors and favors development along the Tcell lineage.
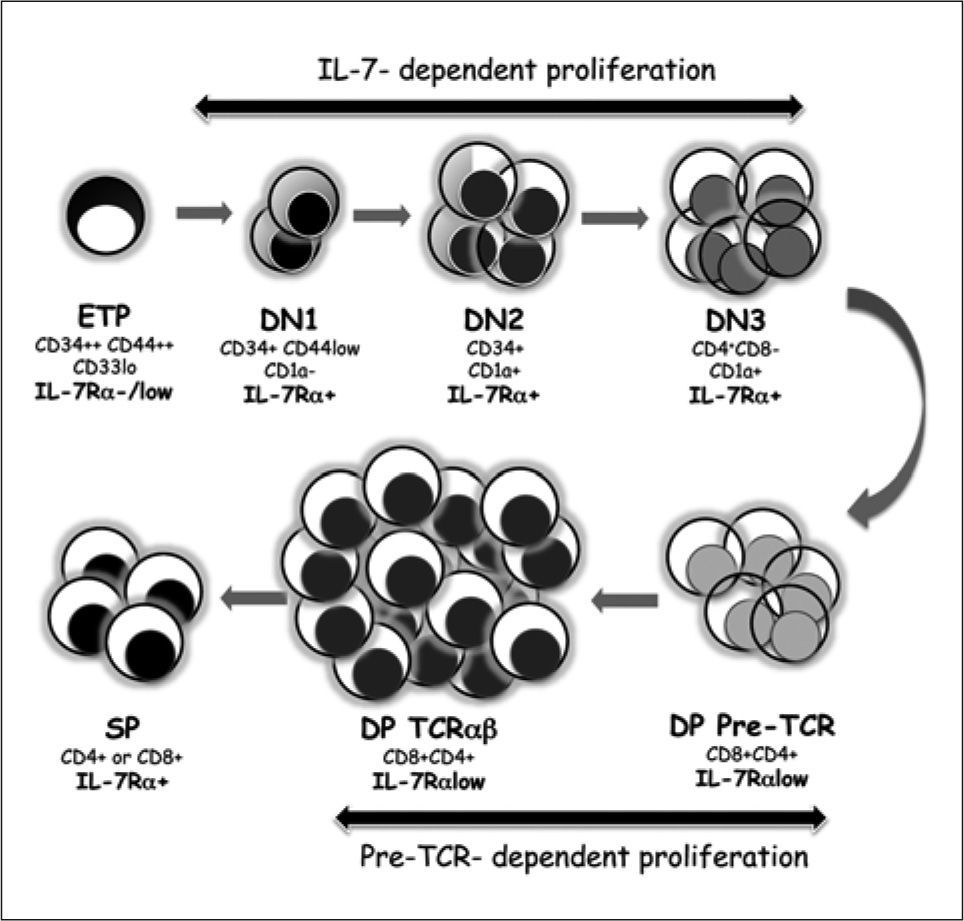
Schematic representation of human intrathymic T-cell developmental stages. Early thymic progenitors (ETPs) progress along double negative (DN) intrathymic stages characterized by the progressive loss of CD34 and CD33 (DN1) and the acquisition of CD1a (DN2) and CD4 molecules (DN3). TCR, gene rearrangements are induced at the DN3 stage, at which TCR, is expressed on the cell surface associated with the invariant pTα chain and CD3 molecules. Pre-TCR signalling induces progression to the double positive (DP) stage (a process known as β-selection) and promotes TCRα gene rearrangements, thus resulting in the generation of TCRaαβ DP thymocytes. TCRαβ-mediated positive and negative selection processes finally result in the generation of MHC-restricted self-tolerant CD4 or CD8 single positive (SP) thymocytes, which migrate to the periphery as mature T cells. Two major waves of proliferation regulated by Notch1 signalling guarantee the generation of the T-cell pool. The IL-7/IL-7R pathway first controls the proliferation of DN T-cell progenitors before β-selection, while later on pre-TCR signaling induces the survival and proliferation of TCRαβ- committed thymocytes.
The hematopoietic system of mammals produces billions of mature blood cells every day in response to physiological demand. HSCs resident in the bone marrow are ultimately responsible for maintaining the lifelong output of new blood cells owing to their pluripotency and extensive self-renewal capability. All blood lineages but T cells derive in situ within the bone marrow from HSCs through a process characterized by the progressive loss of developmental potentials and the activation of lineage-specific transcriptional programs, which ultimately define the specialized function of the mature cell. T lymphocytes, however, develop within the thymic microenvironment from HSCs-derived progenitors that have lost self-renewing capacity. Therefore, the thymus requires periodic or continuous input of hematopoietic progenitors to maintain T cell development. These hematopoietic progenitors must traffic through the circulation from the bone marrow to enter the adult thymus at the corticomedullary junction, and progeny of these entering cells occupy the perimedullary cortical zone, before migrating outward toward the subcapsular zone where substantial proliferation occurs. Thymic progenitors develop through successive developmental stages that are generated in different stromal niches in the thymus owing to cellular interactions with stromal cells, mainly TECs that deliver Notch and other signals that are essential for intrathymic T cell differentiation(1, 2).
The particular identity of the thymus immigrant population has been a matter of debate during the last years, principally because only a few cells are required for efficient thymopoiesis(25). Recent phenotypic and functional studies by different groups, including ours, concurred that the postnatal thymus in both mice and man is seeded by hematopoietic progenitors with a distinctive ckit+Flt3+ IL-7R-/l° phenotype(14, 26, 27), equivalent to that of lymphomyeloid stem cells in the bone marrow(28). In humans, these early thymic progenitors (ETPs) display low CD33 expression levels (CD33lo)(14), while in mouse they express the chemokine receptor CCR9+(29). Such ETPs, which represent 0.01-0.03% of the neonatal thymus, are included among the most immature CD4-CD8- double negative (DN) thymocytes (termed DN1) that display high CD34 and CD44 expression in humans (CD34hi CD44hi)(14, 30) and are CD44+ CD25- in mice(25) (Figure 2). A great advance towards understanding how ETPs undergo T-cell development within the thymus was obtained from initial studies in humans, and later confirmed in mice. These studies showed that such ETPs are multipotent lympho-myeloid progenitors (LMPs) that retain the capability of developing into NK cells, myeloid cells such as macrophages and dendritic cells (DCs), and even give rise to granulocyte-macrophage (GM) colonies, in addition to T-ceUs(14'30-37) (Figure 1). ETPs can also generate B cells(28, 37), although at low frequencies, suggesting that the B-cell potential of thymus immigrants is lost immediately after thymus entry(39). Afterwards, ETPs lose myeloid and NK cell potentials and give rise to T-cell committed thymocytes.
T-cell commitment has been proposed to occur sequentially in the mouse(1-3). In humans, however, progression towards the T- lineage fate involves an early split of ETPs into two alternative developmental pathways that proceed through independent myeloid- or lymphoid-primed intermediate progenitors (Figure 1), characterized by downregulation of CD34 and the simultaneous up or downregulation of CD44, associated with exclusive expression of receptors for either GM-colony-stimulating factor (GM-CSF) or IL-7, respectively(30-32). In addition, upregulation of CD33 and CD5 downregulation also mark differentiation into myeloid-primed progenitors that have lost T-cell potential, but are still able to generate DCs and NK cells, while the reciprocal phenotype defines T/NK bipotential lymphoid progenitors, known as pro-T cells(30, 31) (Figure 1).
Loss of non-T cell potential defines transition to the DN2 (or pre-T cell) stage (CD44+CD25+ in mice) and commitment to the T-cell lineage, a developmental stage marked by acquisition of CD1a in humans(40) (Figure 2?). As the numbers of progenitors that enter the thymus are limited, an enormous expansion takes place during the DN1 and DN2 stages(8). Thereafter, DN2 thymocytes progress to the DN3 stage of CD4+ immature single positive (ISP) thymocytes in humans(41) (CD44-CD25+ in mice), at which cells stop proliferating and rearrangements at the TCRδ, γ and β loci take place(42, 43). Cells that succeed in functional TCRγ and TCRδ rearrangements differentiate into TCRγδ T cells. Alternatively, those cells that express a functional TCRβ chain will accomplish progression beyond the DN3 stage by signalling through a pre-TCR complex composed of TCRβ associated with pTα and CD3 invariant chains, which is highly similar to the murine pre-TCR(20, 44, 45). Pre-TCR signalling promotes survival, proliferation and further differentiation into CD4+CD8+ double positive (DP) thymocytes (DP pre-TCR+) (Figure 2). This process, known as β-selection, involves a great expansion of TCRαβ-committed DP thymocytes that stop cell division after pre-TCR down-regulation(44, 45), undergo rearrangements at the TCRα locus(46, 47) and finally express a mature TCRαβ. Conventional DP TCRαβ+thymocytes will then undergo positive and negative selection processes based on TCRαβ recognition of self-antigens presented by major histocompatibility complex (MHC) molecules expressed by TECs. As a result, TCRαβ+CD4+ or CD8+ single positive (SP) thymocytes are produced(48). Downregulation of CD1 at the SP stage marks acquisition of functional maturation of human thymocytes(49) followed by migration to the periphery as MHC-restricted selftolerant T-cells (Figure 2). Although there is now ample evidence that intrathymic T-cell development is a stepwise Notch-dependent process, the particular role that Notch signalling plays at successive stages of early thymopoiesis is still not fully understood and current knowledge on this issue is described below.
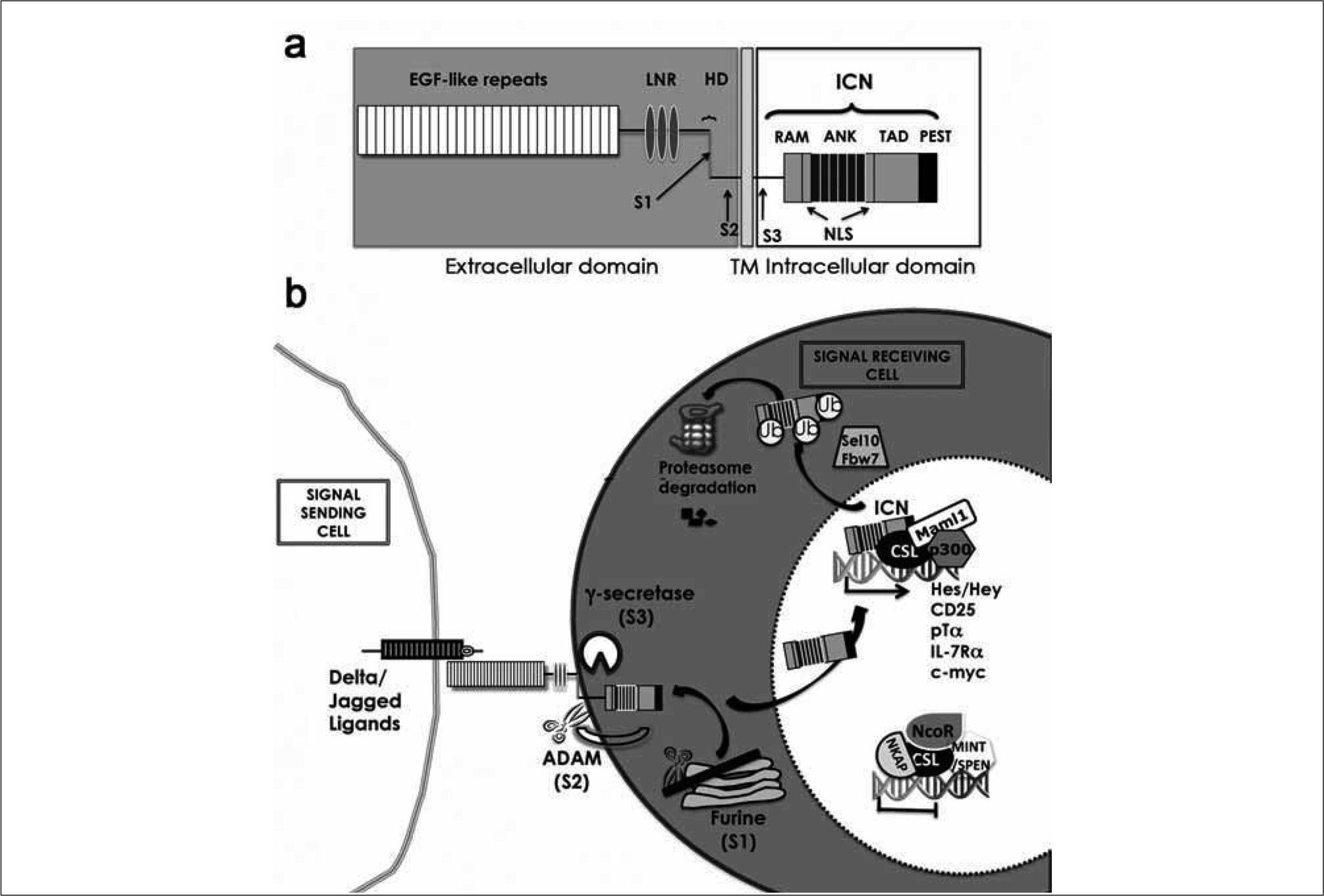
NOTCH STRUCTURE AND SIGNALLINGNotch receptors represent a highly conserved family of transmembrane proteins(50) comprising four members (Notch1-4) in mammals. Notch receptors are synthesized as a 300kDa precursor that is fucosylated in the endoplasmic reticulum and is cleaved at the S1 site in the trans-Golgi network by a furine-like convertase. The resulting fragments are non-covalently associated into a mature heterodimer receptor that is exported to the cell surface(51). The extracellular (EC) domain of Notch contains 36-tandem epidermal growth factor (EGF)-like repeats, some of which mediate interactions with the Notch ligand (Figure 3a). The EC domain is followed by a unique negative regulatory region (NRR) composed of three Lin 12-Notch repeats (LNR) and a heterodimerization domain (HD), responsible for stable subunit association, which maintains the receptor in an inactive conformation in the absence of ligand(52). Intracellularly, Notch contains a RAM (RBPj¿ association module) domain, a number of nuclear localization sequences (NLS), seven cdc10/ankyrin repeats (ANK domain) and a transactivation domain (TAD) harbouring a PEST sequence(52). The RAM sequence is required for interaction with the transcription factor CSL (CBFl/RBPJ¿ in mammalian cells, Su(H) in Drosophila and LAG-1 in C. elegans) and further regulation of target gene transcription(52). ANK repeats associate with coactivators of the Mastermind-like family (MAML-1-3), which are obligatory for Notch signalling(53, 54), while the PEST domain regulates protein stability through targeting by ubiquitin ligases required for proteosome-dependent Notch degradation and thereby for termination of signalling(55) (Figure 3a).
Structure and function of Notch receptors. (A) The extracellular domain of Notch receptors contains 36-tandem epidermal growth factor (EGF)- like repeats involved in the interaction with Delta/Jagged ligands expressed on neighbor cells, a negative regulatory region containing Lin 12-Notch repeats (LNR) and a heterodimerization domain (HD). The intracellular domain (ICN) contains a RBP-Jk association module (RAM), ankyrin repeats (ANK), a transactivation domain (TAD) and the PEST domain, implicated in ICN degradation. Cleavage sites for furine-like convertases (S1), ADAM metalloproteases (S2) and the γ-secretase complex (S3) are indicated. (B) Upon interaction of Notch receptors with their ligands, two proteolytic cleavages (S2, ADAM; and S3, γ-secretase) release the ICN domain from the membrane. ICN then migrates to the nucleus where it displaces co-repressors, including NCoR, NKAP and MINT/SPEN, from the CSL transcription factor and recruits co-activators, such as MAML-1 and p300/CBP, to finally induce the transcription of target genes, including HES, HEY, CD25, PTCRA and IL7R. Termination of Notch signalling is induced following ubiquitination by E3 ubiquitin ligase Sel-10/Fbw7 and proteasome degradation.
Notch receptors recognize cell surface ligands of the Delta-like (DL1, DL3 and DL4) and Jagged (Jag l and Jag 2) families(56) expressed on neighbor cells (Figure 3b). Ligand affinity can be modulated by glycosylation of O-linked fucose residues attached to EGF-repeats by the Fringe family of glycosyltransferases, specifically Lunatic Fringe in T-cell development(57). Ligand interaction forces conformational changes in the Notch receptor that expose the cleavage site (S2 site) for ADAM (a disintegrin and a metalloprotease) metalloproteases, specifically ADAM10 (Kuzbanian) and ADAM17 (TNF-α converting enzyme, or TACE) members, leading to the generation of a membrane-anchored Notch isoform lacking the extracellular region (Figure 3b). The shedding of extracellular Notch creates the substrate for a γ secretase protein complex (presenilin, APH-1, nicastrin and PEN-2), which proteolytically cleaves the receptor at the yuxtamembrane region (S3 site), releasing intracellular Notch domain (ICN or NICD) that migrates to the nucleus and associates to CSL(52). ICN binding to CSL displaces co-repressors and recruits co-activators including p300/CBP and MAML-1-3, thereafter initiating transcription of target genes. Canonical Notch targets are basic-helix-loop-helix proteins (bHLH) such as Hey and Hes(58). Other Notch targets include genes involved in cell proliferation and survival such as c-myc(59), cyclin D1(60) and p21/Waf(61). In addition, CD25(62) and pTα(63) are known targets of Notch in T cells (Figure 3b).
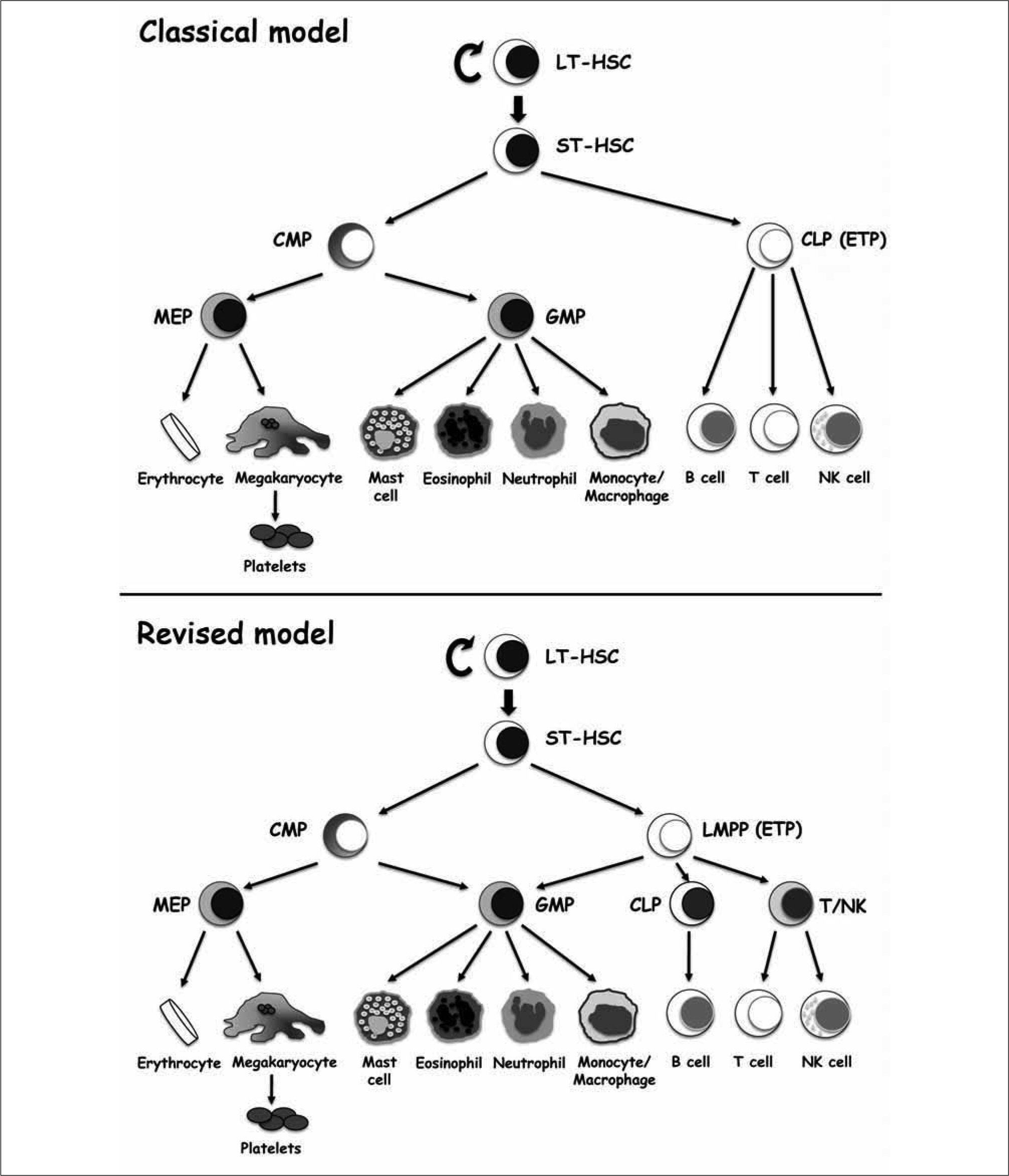
NOTCH IN T CELL DEVELOPMENTThe critical role of Notch in the development of T cells was first described by Ratdke and coworkers(6) helped by the generation of inducible loss-of-Notch1 function (Notch1−/−) mice. Notch l-deficient mice showed a smaller thymus with a 5-fold reduction in total thymocyte numbers and an abnormal architecture of both cortical and medullar thymic regions. T-cell development in those mice was blocked at the most immature T-cell stage, while B220+ B cells were 200-fold increased in the thymus. Complementary to these findings, gain-of-function approaches showed that Notch1 was sufficient to promote extrathymic T-cell development(7). When bone marrow HSCs cells genetically modified to express constitutively active ICN1 were transferred into irradiated hosts, a thymus-independent DP T-cell population appeared in the bone marrow and B cell differentiation was completely inhibited. The conclusion of these studies was that Notch1 signalling critically influences B versus T lineage choice during hematopoiesis, driving T/B common lymphoid progenitors (CLP) towards the T lineage. However, the CLP concept(64) has recently been challenged by the emergence of revisited models of hematopoiesis(65, 66) supporting a closer relationship between lymphoid and myeloid lineages than "conventional" models(67) (Figure 4) and, ultimately, by the finding that lymphoid-primed multipotent lympho-myeloid progenitors, rather than CLPs, include the canonical T-cell precursors in the postnatal thymus(13, 27-39, 68-71). Accordingly, the initial proposal on the role that Notch signalling plays in early thymopoiesis was re-examined.
Classical and alternative models for adult hematopoietic stem cell lineage commitment. Classical model for hematopoietic development postulates that lineage commitment of long-term hematopoietic stem cells (LT-HSCs) underlies an strict separation of myelopoiesis and lymphopoiesis that involve independent CMP and CLP progenitors, respectively. The alternative model suggests that HSCs give rise to a lymphoid-primed intermediate progenitors that retain myeloid potential upon loss of Mk and E potential. Generation of T cells from these progenitors occurs upon loss of B cell potential and independent generation of CLPs. LT-HSC, long-term hematopoietic stem cell; ST-HSC, short term HSC; LMPP, lymphoid-primed multipotent progenitor; CMP, commom myeloid progenitor; MEP, megakaryocyte/erythroid progenitor; GMP, granulocyte/monocyte progenitor; CLP, commom lymphoid progenitor; ETP, early thymic progenitor.
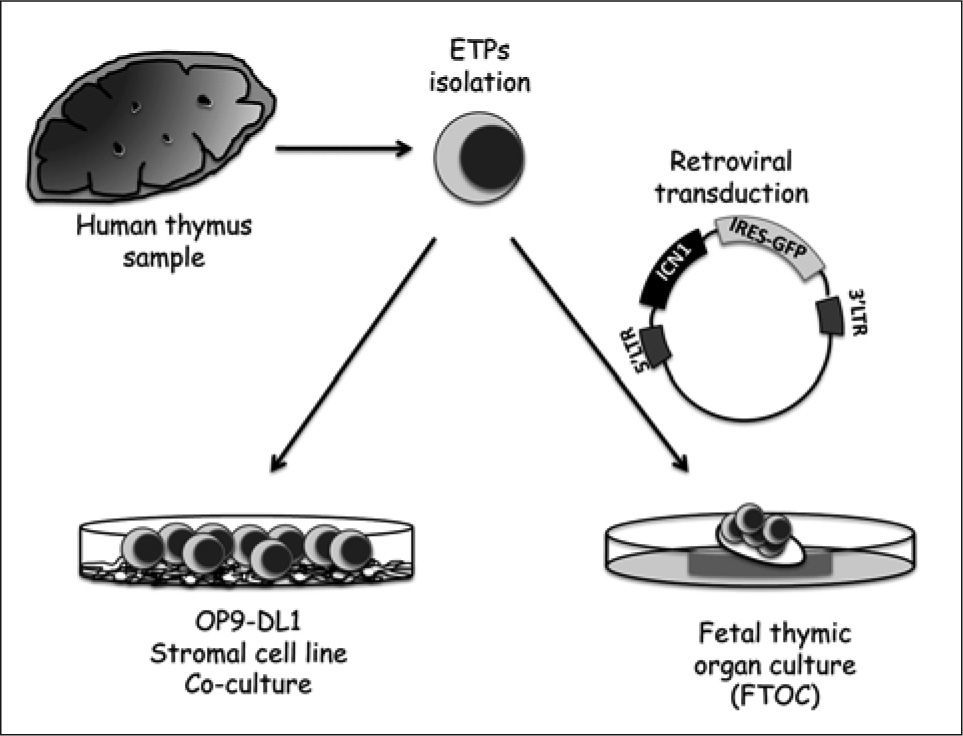
Regarding the role of Notch in humans, we and others took advantage of gain-of-function approaches aimed at inducing either ligand-independent Notch1 signalling in ETPs by ectopic expression of active ICN1(14, 68), or liganddependent Notch1 activation(13, 14) by co-culturing ETPs onto OP9 stromal cells expressing the Notch ligand Delta-like1 (OP9-DL1), as described by Zúñiga-Pflücker in mice(72) (Figure 5). Both strategies provided evidence that the most prominent role of Notch1 signalling in early human T-cell development is to block non-T cell differentiation (Figure 1). Particularly, Notch1 activation is able to impair the differentiation of ETPs into myeloid-primed intermediate progenitors(14), thereby inhibiting the generation of DCs and macrophages, and it also blocks the development of NK cells from T/NK bipotent progenitors(13, 14). Inhibition of non T cell fates by active Notch1 promotes the exclusive development of T-lineage cells and results in the transcriptional induction of specific T-lineage genes(1-3, 63), a finding that supports an instructive role of Notch1 in T-cell specification. However, while Notch activation itself is sufficient to block non-T cell development, additional stromal-derived inductive signals are concurrently required to promote full differentiation along the T-cell lineage. In fact, ETPs over-expressing ICN1 are unable to undergo TCR rearrangements and to progress beyond the DN1 developmental stage, unless co-cultured with stromal cells(14). Such ICN1+ DN1-like progenitors display a great proliferation capacity, a feature also observed in HSCs from cord blood following Notch1 activation(73, 74). Thus, Notch signalling was proposed to play a key role in the maintenance and expansion of HSCs and thymic progenitors. Importantly, Notch-induced cell proliferation of both HSCs and ETPs was found dependent on unique signals provided by cytokines (see below).
Experimental strategies used to analyse the impact of Notch1 signalling in human T-cell differentiation. Different studies have focussed on the impact of either ligand-dependent or ligand-independent Notch signalling on the developmental potential of human ETPs. For liganddependent Notch activation, ETPs are co-cultured onto OP9 stromal cells constitutively expressing the Notch ligand Delta-like 1 (OP9-DL1). Ligandindependent Notch signalling is induced by retroviral transduction with a bicistronic vector encoding active ICN1 and green-fluorescent protein (GFP) as cell tracer. Differentiation of transduced cells is then assessed in a human/mouse fetal thymic organ culture (FTOC) assay.
Complementary loss-of-function approaches have been also designed in an effort to address the consequences of Notch signalling inhibition and to better understand the role of this pathway in early T-cell development. Gammasecretase inhibitors (GSI), small pharmacological molecules that block Notch cleavage(75), have been particularly useful as inhibitors of Notch signalling in vitro. In addition, Notch inhibition has been accomplished in vivo by engineering a dominant negative construct of the MAML-1 co-activator (DNMAML-1) that binds the CSL/Notch complex and blocks Notch-dependent transcription(76). These strategies supported the proposed regulatory function of Notch1 in the amplification of the intrathymic pool of T-cell progenitors(14, 24). However, recent in vivo studies on Notch inhibition in mouse ruled out an essential physiological role for canonical Notch signals in maintenance and expansion of more immature HSCs(77), despite pioneering gain-of-function studies by Irvin Bernstein and coworkers supported the involvement of Notch signalling in self-renewal of HSCs(78). Whether these discrepancies rely on the experimental strategy used, or reflect a differential contribution of Notch to hematopoiesis under steady state or stress conditions remains to be clarified.
Loss-of-function studies also confirmed that Notch signalling is essential to impair intrathymic differentiation of non-T lineage cells. Using increasing concentrations of GSI, Plum and coworkers concluded that gradual loss of non-T cell differentiation options induced by Notch1 can be explained in quantitative terms(13), since increasing thresholds of Notch signalling sequentially suppress B, myeloid/DC and NK cell lineage fates in humans, as suggested in mice(12). How thus is Notch signalling regulated in quantitative terms?. The answer to this question comes from pioneering studies by Parreira and coworkers in humans(79), later confirmed in mice(80), showing that different Notch ligands transmit distinct activation signals to T-cell precursors that differentially affect their proliferation and/or differentiation potential. Particularly, Jagged ligands seem to induce lower Notch activation in comparison with DL ligands(57), suggesting that expression of distinct Notch ligands at particular intrathymic locations could be responsible for the existence of different Notch activation thresholds along thymopoiesis. However, formal proof was recently provided that DL4 is the essential, nonredundant ligand for Notch1 during thymic T-cell lineage commitment(81) and, therefore, alternative mechanisms such as differential regulation of DL4 expression levels may underlie the quantitative control of Notch signalling within the thymus.
Quantitative Notch1 signals have been also suggested to influence the TCRα, versus TCRγδ decision(82), and recent data indicate that Notch1 might mediate this event by synergizing with signals generated through the TCR(83, 84), although contradictory results have been obtained in mice and man. In the mouse, TCRγδ development is less Notch dependent compared with TCRα, differentiation(82, 84), a finding that concurs with a higher dependency of the former on Jagged ligands(85). In contrast, sustained Notch1 signalling favours TCRγδ development at the expense of TCRα, generation in Fetal Thymic Organ Culture (FTOC) in humans(68), and similar results have been described using OP-DLl cocultures(86). While these data suggest that an early decrease in Notch1 activation is required for human TCRα, lineage differentiation at the expense of TCRγδ T cells (86), our recent studies using xenotransplanted Rag2−/− x γcr−/−/immunodeficient mice indicate that ICN1 over-expression does not affect the final outcome of human T-cells in vivo (unpublished results). Therefore, the influence of Notch1 signalling in the regulation of binary cell fate decisions at the critical α, versus γδ branching point remains unclear in humans, although Notch is certainly required earlier in thymopoiesis for the generation of both cell types(13, 14).
Notch signalling also participates downstream of T-cell commitment at the critical β-selection check-point. Human ETPs are unable to fully differentiate into mature DP TCRαβ+cells in GSI-treated FTOC(13, 68, 77) or OP9-DL1 co-cultures(14). Instead, an aberrant DP population lacking CD3 and cytoplasmic TCRβ (TCRβ ic) is generated, which is equivalent to that observed in Notch1-deficient mice(87), and high frequencies of NK cells, monocytes and DCs are also produced. Therefore, Notch1 signals are required not only to block diversion away from the T-cell fate, but also to complete Tcell maturation of TCRαβ -committed progenitors. While these results are compatible with an impaired TCRβ gene rearrangement in the absence of Notch activation, recent data by Maillard and coworkers in mice(22), point toward an absolute requirement of Notch signalling for cell survival/proliferation during β-selection in vivo, which seems pre-TCR-independent, as shown before in vitro(21). It is thus believed that Notch and pre-TCR act in parallel pathways that may synergize during β-selection to maintain proliferation and survival both in mice and humans(l4,68,88,89). Finally, although Notch1 was initially proposed to participate in the CD4 versus CD8 lineage decision during positive selection, this conclusion was later challenged by studies using mice made conditionally deficient for Notch1, which formally excluded an essential role for Notch1 in CD4/CD8 lineage commitment, maturation or survival(90). However, it is becoming increasingly clear that, once in the periphery, cell-fate and functional differentiation of T cells might critically depend on Notch signals(9l). Notch has been proposed to influence Th1 versus Th2 differentiation of CD4+ T helper subsets and Th17 generation as well as activation and proliferation of peripheral T cells, leading to cytokine production or maturation of CD8+ naive T cells into cytotoxic T-lymphocytes (CTL)(9l). However, the field remains controversial and more studies, including genetic approaches in mice made deficient for the different Notch ligands, as well as identification of critical effectors of the Notch pathway, are needed to reach a general conclusion about the role of Notch signalling in those processes.
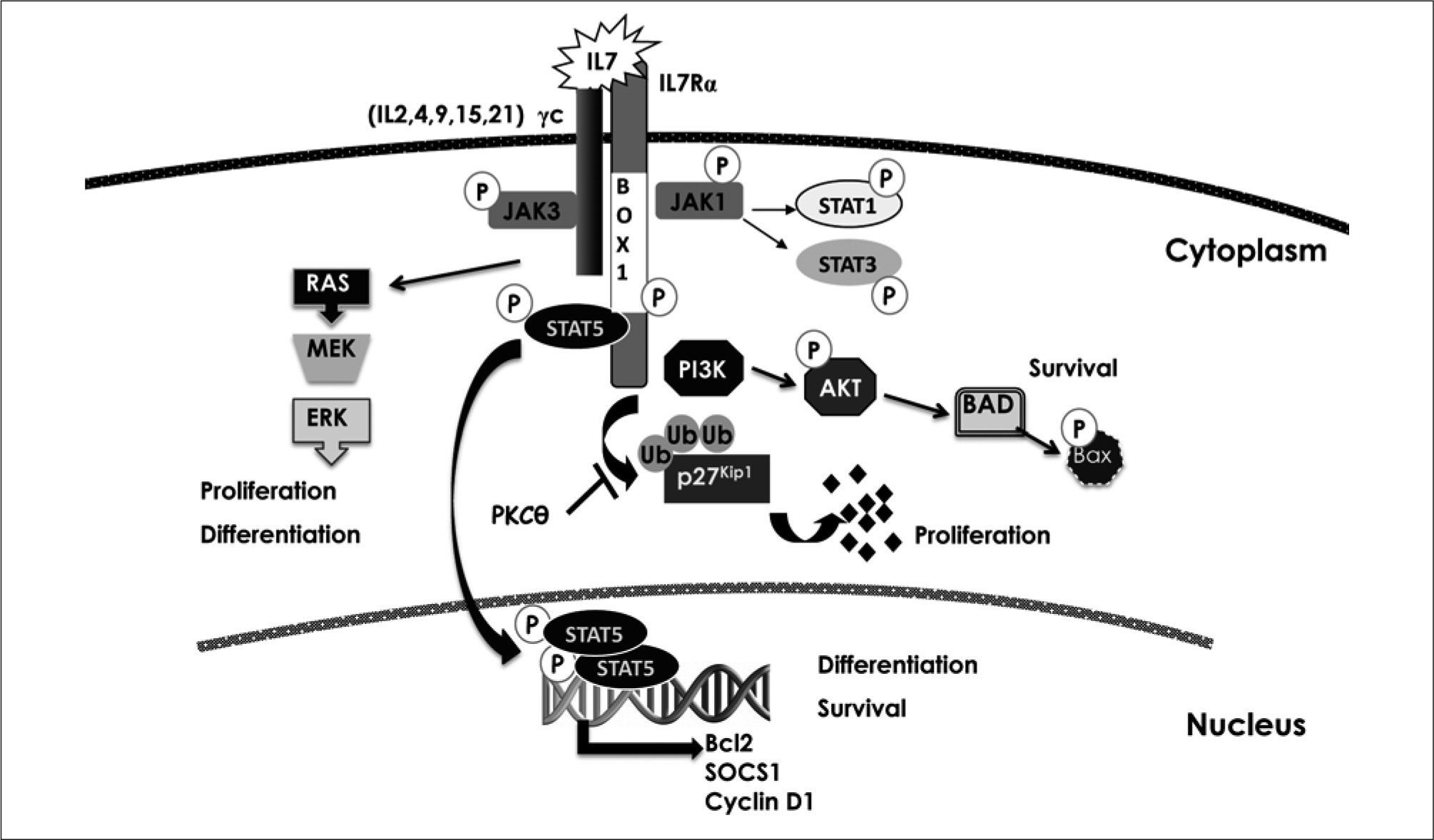
INTERLEUKIN-7/INTERLEUKIN-7 RECEPTOR SIGNALLING IN EARLY T-CELL DEVELOPMENTAs described above, two main phases of cellular proliferation occur after thymus settling to generate the abundant population of DP thymocytes, one before and the other after induction of TCR gene rearrangements (Figure 2). While the latter is controlled by the pre-TCR at the β selection checkpoint, the former is critically dependent upon and mediated by the high affinity receptor for IL-7 (IL-7R)(8, 9, 92-94). IL-7 is a 25kDa cytokine produced by stromal cells within the bone marrow and the thymus that transmits signals for survival and proliferation to T and B cells. Binding of IL-7 to its receptor (Figure 6), which is composed of a specific β-chain (IL-7Rα) associated to the common cytokine receptor γ (γc) chain(95), plays a conserved non-redundant role in thymopoiesis by supporting the survival and expansion of early DN precursors(8, 9, 92-96). However, IL-7 is dispensable beyond the DN3 stage, although it seems to be required later on during positive selection of CD8+ thymocytes(97, 100).
Interleukin 7 receptor signaling pathway. Binding of interleutín 7 (IL-7) to its receptor, composed of an IL-7Rα and a γ-common (γc) chain, promotes receptor dimerization, which results in the activation of JAK tínases bound to the intracellular domain of γc (JAK3) or α-chain (JAK1). JAK-induced phosphorilation of the IL-7Rα intracellular domain creates a docking site for recruitment of SH-2 domain-containing proteins such as STATs and PI3K. Phosphorilation of STATs by JAKs, results in dimerization, migration to the nucleus, and transcription of target genes such as BCL-2, SOCS1 and CyclinD1. PI3K recruitment to the IL-7Rα intracellular domain induces its activation and results in AKT phosphorylation and degradation of pro-apoptotic proteins such as Bad and Bax. MEK1/ERK1/2 mitogen activated protein kinases (MAPK) are also activated by IL-7R signalling, leading to proliferative and differentiation signals. Cell cycle induction is mediated by degradation of the cell cycle inhibitor p27Kip1, in a PKC-dependent fashion.
The stage-specific function of IL-7 during intrathymic development is accomplished by a dynamic regulation of IL-7R expression and function(97). Of the two IL-7R components, IL-7Rγ is constitutively expressed in thymus-seeding precursors, while IL-7Rα is induced at the ETP stage and increases progressively until the DN2/DN3 stage(14, 26, 27). Thereafter, IL-7Rα expression declines steadily(9, 14, 26, 97) and must be terminated between the β-selection and positive selection checkpoints to allow for transition to the DP stage(98-100). To further guarantee IL-7R-unresponsiveness, obligatory for development of DP thymocytes(98), IL-7R signalling is actively suppressed in pre-selection DP thymocytes by suppressor of cytokine signalling (SOCS)-1, but it is restored by positive selection(100). Moreover, down-regulation of IL-7R expression during thymopoiesis is required to avoid a direct competition between DN progenitors and DP thymocytes for the limiting amounts of endogenous IL-7 within the thymus(97). Restoration of IL-7R surface expression by positive selection will finally ensure homeostatic proliferation of peripheral mature T cells derived from CD4 and CD8 SP thymocytes(101).
According to a key role of IL-7R signalling in T-cell development, defective IL-7Rα expression results in a severe combined immunodeficiency syndrome (SCID) in humans characterized by a complete lack of T cells. In mouse, however, both T and B lymphopoiesis are severely affected in the absence of IL-7Rα(93-96), indicating that differential IL-7R signal requirements may exist in mice and humans. Regarding the function of IL-7R in T and B cells, several reports support the existence of linage-specific signalling pathways downstream of IL-7R. Binding of IL-7 to its receptor results in α and γ chain dimerization at the cell surface, which triggers downstream activation pathways including JAK/STAT, PI3K, MAPK and Src kinases, ultimately leading to the regulation of target genes such Bcl-2 family members, cyclin Dl, SOCS-l, c-myc and IL-2Rα (95,l02) (Figure 6). Of them, Bcl-2 is sufficient to rescue T cell development in IL-7Rα- deficient mice (l03,l04), but Bcl-2 cannot rescue impaired B lymphopoiesis in those mice(l05). These results point toward the pro-survival function as the most important function of IL-7 during T-cell development. Interestingly, PI3K is the specific pathway controlling survival and proliferation of T cell precursors, while STAT5 activity is linked to T-cell differentiation(l06) and also controls rearrangements at the TCRγ(l07) and IgH gene loci(l08) in T-cells and B-cells, respectively.
NOTCH1 SIGNALLING CONTROLS T LINEAGE SPECIFIC EXPRESSION OF IL-7R IN EARLY THYMOPOIESISThe stage- and lineage-specific functions reported for IL-7 indicate that strict mechanisms may control the dynamic regulation of IL-7R expression in the thymus and its differential expression in T- and B-lineage cells during lymphopoiesis. It is known that murine IL-7Rα gene (Il7ra) transcription in early lymphoid and B cell progenitors is specifically regulated by the Ets transcription factor PU.l(l09). PU.l is also expressed very early in thymopoiesis in ETPs, but it must be obligatory down-regulated for progression along the T-cell lineage(110). Then, the Ets transcription factor GA binding protein (GABP) seems to replace PU.l for controlling IL-7Rα expression in mature T-cells, although the involvement of GABP during early thymopoiesis is less clear(111). In contrast, GABP cooperates with PU.l in the B-cell lineage and regulates IL-7Rα expression in pre-B and committed B-cells(112). While these data provide direct evidence for the existence of specific regulators of IL-7Rα expression in B cell development, the nature of the equivalent regulators in the T-cell lineage has remained an open question until very recently, when results from our laboratory demonstrated that Notch1 accomplishes this function in humans(24). By inducing ligand-dependent (co-culture onto OP9-DL1 stroma) or independent (ICNl retroviral over-expression) Notch1 activation, we found that Notch1 signalling is required for IL-7Rα expression in the T-cell progeny arising from either human HSC or ETP progenitors(68, 77). In fact, Notch1 signalling specifically promotes IL-7Rα gene (IL7R) transcription in human T-, but not B-lineage cells, which can be inhibited by ectopic expression of DNMALM-1. Formal proof that Notch1 directly controls IL7R gene expression in developing T cells was provided by luciferasereporter and chromatine immunoprecipitation assays showing that IL7R is indeed a downstream target of Notch1 in human thymocytes(76). Supporting a CSL-dependent mechanism of Notch1-induced IL7R gene activation, we have identified a putative CSL-binding site in the human IL7R promoter that is conserved in mouse and have shown that either site-directed mutagenesis at the CSL-binding site or CSL-deficiency (in RBP-j¿-deficient mice) impairs Notch1-induced IL7R promoter activity. Therefore, CSLMALM-dependent Notch1 signalling controls T-lineagespecific IL7R gene expression in humans(24). Confirming a fundamental role of Notch1 in the regulation of IL-7Rα in human thymopoiesis, Notch activity parallels IL7R transcription levels and mRNA expression of BCL2 and PTCRA Notch1 targets along human T cell development in vivo. Moreover, defective Notch1 signalling selectively results in a compromised expansion of the early pool of thymocyte precursors, which can be rescued by ectopic IL7Rα expression. This is a stage-specific effect restricted to thymocytes within the early DN compartment, but IL-7R signalling is unable to replace Notch1 signals required in combination with pre-TCR signals at the β-selection checkpoint(24). Collectively, these results demonstrate that Notch1 signalling controls proliferation of T cell progenitors upstream of β-selection by regulating the lineage- and stage-specific expression of IL-7Rα, thus providing competence to developing T cells for responding to local supply of IL-7.
NOTCH1 SIGNALLING IN THE PATHOGENESIS OF T-CELL LEUKEMIAT-acute lymphoblastic leukemia (T-ALL) is a lymphoproliferative disorder accounting for l0 to l5% of pediatric and 25% of adult ALL cases (113) that results from the malignant transformation of normal developing T cells in the thymus(114, 115). Aberrant Notch1 signalling was initially described in human T-ALLs (<l%) with rare chromosomal translocations that generate the expression of a truncated Notch1 isoform lacking the extracellular domain (TANl) under the transcriptional control of the TCRβ enhancer(115). Studies that are more recent revealed that activating mutations of Notch1 are common in more than 50% of human TALLs(23), this finding supporting a role for Notch1 signalling in leukemogenesis more prominent than initially suspected. Activating Notch1 mutations in T-ALLs are mainly located in the HD and/or in the intracellular PEST domains (Figure 3a). HD mutations expose the S2 site of Notch1, increasing ADAM cleavage and subsequent receptor activation in the absence of ligands. PEST mutations are short insertions or deletions that result in partial or complete absence of PEST domain, thus increasing Notch1 stability and half-life(23).
T-ALLs, initially linked to a very poor prognosis, have shown an improved outcome in recent years due to intense chemotherapy treatments. Still, patients with relapsed disease (15-40% and 40-50% in paediatric and adult cases, respectively) continue to have a bad prognosis. Investigators have therefore focused on understanding the molecular mechanisms downstream of Notch1 responsible for disease induction and maintenance, with the final aim of identifying new therapeutic targets for T-ALL based on Notch1 inhibition. Several Notch-related molecular pathways involved in normal T-cell development have been implicated in Tcell transformation. Initial studies showed that mice transplanted with bone marrow progenitors expressing active Notch1 alleles rapidly develop aggressive T cell leukemias(7, 118), but only when signalling mediated by the pre-TCR complex was intact(119). These results supported a synergistic role between Notch1 and pre-TCR signalling in leukemogenesis, as also shown for the Notch3 receptor(120). Whether the two pathways act in a linear or in a parallel way remains an open question. Supporting the first possibility, it was shown that the PTCRA gene is a transcriptional target of Notch activity(121), and expression of other pre-TCR components such as CD3ε and TCRβ are also controlled by Notch(114). It is therefore possible that the Notch pathway could be upstream of pre-TCR expression. An alternative possibility is that Notch1 and pre-TCR pathways act in parallel but converge at signalling intermediates in T-ALLs, as both pathways can activate the transcription factors cmyc, NF-¿B and NFAT and the kinases LCK, IKK and the PI3K-AKT pathway and share common target genes such as cyclin D3 and BCL-2A1(114). According to this second view, c-myc, which is a crucial regulator of cellular metabolism and cell cycle progression, has been identified as a key target in Notch1-dependent leukemogenesis(59, 121). In fact, conditional ablation of c-myc in ICN1-expressing DP cells prevents tumour formation, indicating that both ICN1 and c-myc are required for tumour maintenance(122). Similarly, Notch1 signalling leads to activation of the NF-¿B pathway, which seems to be a downstream target of Notch1 in TALL(123). In addition, Notch1 has recently emerged as a negative regulator of PTEN in T-ALLs(124). Notch1-dependent Hes1 expression controls the levels of PTEN, thus affecting the balance between activation/inhibition of PI3K, one of the major inducers of proliferation and survival in T cells. Moreover, PTEN inactivating mutations and constitutive PI3K activation have been identified in Notch1-dependent T-ALLs. In this scenario, Notch1 inhibition was not sufficient to control PI3K activity and cells were able to proliferate independently of Notch1 by constitutive activation of PI3K and AKT(124). Therefore, although therapeutic strategies directed to Notch1 inhibition, particularly treatment with GSI, initially emerged as a promising therapy owing to its ability to decrease Notch1 expression and leukemic cell viability(23), our more recent knowledge of the molecular pathology of the disease demands the design of novel targeted therapies.
NOTCH1 AND IL-7R INTERPLAY IN T-CELL LEUKEMIAOur recent identification of IL7R as a downstream target of Notch activity points toward the IL-7/IL-7R pathway as a potential candidate to induce or maintain T cell leukemogenesis. This appears a likely possibility considering the prominent role that IL-7 plays in T-cell survival and proliferation and, specifically, because human T-ALLs express functional IL-7 receptors that respond to exogenous IL-7 and significantly contribute to T-ALL proliferation by activation of PI3K(125). Accordingly, we have shown that IL-7Rα expression is specifically downregulated when Notch1 signalling is inhibited by GSI treatment or DNMAML-1 ectopic expression in TALL, but not in B-ALL, cell lines, this resulting in IL-7- unresponsiveness, as judged by defective STAT5 and AKT phosphorylation. More importantly, impaired IL-7R signalling in T-ALLs leads to decreased proliferation and cell cycle arrest, which can be rescued by ectopic IL-7Rα expression and results in a selective growth advantage of IL-7Rα-expressing T-ALL cells(24). Overall, our study indicates that IL-7/IL-7R signalling is able to support the survival and expansion of leukemic cells with impaired Notch signalling, and points toward IL-7Rα as a major regulator for cell cycle progression induced by Notch1 in T-cell leukemia. It is therefore essential to develop new therapies that target oncogenic pathways in Notch1 dependent T-ALLs, such as the IL-7R pathway.
CONCLUSIONS AND REMARKSThe study of the molecular mechanisms regulated by Notch1 activation in human T-cell development and T-cell leukemias higomplex in response to Notch1. Therefore, understanding Notch1 signalling effectors involved in physiologic and oncogenic proliferation is crucial for designing new therapeutic strategies that target relevant oncogenic pathways in T-ALLs. The identification of IL-7Rα as a downstream target of Notch1, both in physiology and pathology, together with the fihlights the close relationship between Notchdependent signalling pathways in T-cell physiology and pathology. Indeed, the most prevalent function of Notch1 in T-cell development is the induction of cellular expansion: first, expansion of the pool of early T-cell progenitors that have been devoid of non-T cell potentials in response to Notch1, and thereafter expansion of those progenitors that successfully progress along the T-cell maturation pathway and express a functional TCRβ-pTα pre-TCR signalling cnding that maintenance and expansion of Notch1-dependent TALL leukemic cells can be modulated by manipulation of IL-7R signalling, open new possibilities to develop specific targeted therapies. Preclinical studies exploring the efficiency of IL-7R targeted therapies in xenograft models of Rag2 x γc-deficient mice transplanted with human T-ALL cells are presently underway and offer promise for more effective TALL therapies in the near future.
CONFLICT OF INTERESTThe authors declare no financial conflict of interest.
This work was supported by grants from Plan Nacional (BFU 2007–60990), Comunidad de Madrid (S-SAL0304-2006), Fundación la Caixa (ON03/l09-00), Fundación MM, and Instituto de Salud Carlos III (RECAVA RD06/00l4/l0l2) and by an Institutional Grant from the Fundación Ramón Areces. S.G-G was supported by Ministerio de Ciencia e Innovación (MICINN) (FPI program). The authors declare no conflicting financial interests.