Neuronal function and differentiation are tightly regulated by both genome and epigenome. Based on the environmental information the epigenetic changes occur. Neurodegeneration is the consequence of dysregulation of both the genome and epigenome. In this study, we saw different types of alterations of epigenome present in neuronal cells of different model organisms for neurodegenerative disorders. The epigenetic modifications including chromatin modification, DNA methylation, and changes in regulatory RNAs (miRNA) are having a great impact on neurodegenerative disorders as well as memory. The effects of these re-editing in the neuronal cells cause Alzheimer's disease, Parkinson's disease, Huntington's disease but an unusual form of neuroepigenetics has been seen in Prion Disease. Subsequently, for the development of treatment of these diseases, epigenetic modifications should be kept in mind. Although until now many reports came on drug discovery inhibiting histone deacetylases and DNA methyltransferases to reverse the epigenetic change but they lack targeted delivery and sometimes cause a cytotoxic effect on neuronal cells. In future, advancement in targeted and non-cytotoxic drugs should be the main focus for therapeutic treatment of the neurodegenerative disorders.
La función y diferenciación neuronales están reguladas en gran medida por el genoma y el epigenoma. Los estímulos ambientales producen cambios epigenéticos. La neurodegeneración es consecuencia de una alteración en el genoma y el epigenoma. Hemos analizado diferentes tipos de alteraciones del epigenoma presentes en células neuronales de diferentes modelos animales de enfermedad neurodegenerativa. Los cambios epigenéticos (modificación de la cromatina, metilación del ADN, cambios en los ARN reguladores [miARN]) tienen un impacto importante en las enfermedades neurodegenerativas y en la memoria. Dichos cambios en células neuronales causan diferentes enfermedades, como las de Alzheimer, Parkinson, y Huntington; sin embargo, las enfermedades priónicas muestran formas epigenéticas inusuales. Por tanto, el desarrollo de tratamientos para estas enfermedades debe considerar los cambios epigenéticos. Se han desarrollado diversos fármacos inhibidores de la histona deacetilasa y la ADN metiltransferasa, que revierten los cambios epigenéticos, pero no utilizan sistemas de liberación inteligente, por lo que a veces pueden producir efectos citotóxicos en las células neuronales. La investigación sobre tratamientos para las enfermedades neurodegenerativas debe centrarse en el desarrollo de fármacos no citotóxicos con sistemas de liberación inteligente.
Epigenetics is referred to as meiotically and/or mitotically heritable changes in gene expression which are not coded by nucleotide sequences in DNA but by interoceptive as well as by the influence of environmental factors. Histone modifications, DNA methylation and alteration in both coding and non-coding RNAs resulting in the changes in the expression of genome are part of epigenetic modifications.1 All these modifications have role to play in gene and protein expression without alteration of DNA sequences. Catastrophic repercussion in neural cells degeneration might be caused by the epigenetic modification.
Neurodegenerative diseases like Alzheimer's disease (AD), Parkinson's disease (PD), Huntington's disease (HD), Prion disease and others are greatly influenced by environmental factors therefore changes in epigenome are involved. The epigenetic related studies in neurons are termed as neuroepigenetics. Unlike classical epigenetics neuroepigenetic modifications are not inherited. For information storage and circuit regulation epigenetic mechanisms play an enormous role.2 Neuroepigenetics can interfere with both development and function of central nervous system. An exponential expansion is seen in this field over 25 years. In this review we mainly focus on the neurodegenerative disorders as a consequence of epigenetic modifications.
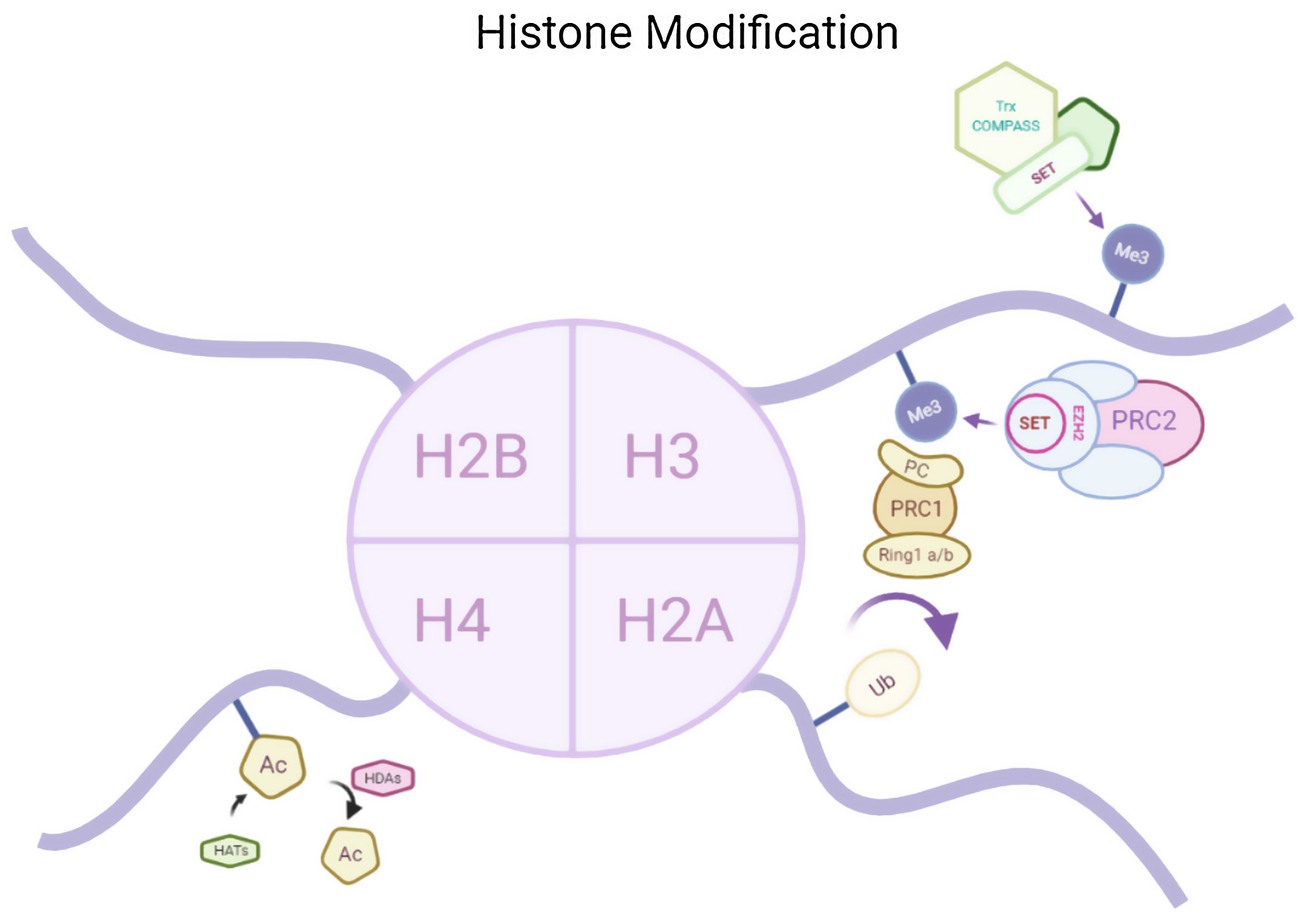
Epigenetics and memoryHistone acetylationHistone acetylation brought by histone acetyl transferases (HATs) activates the transcription by formation of euchromatin structure whereas histone deacetylase (HDAC) causes deacetylation therefore causing transcription repression by formation of heterochromatin structure. Both of the two enzyme are responsible for synaptic plasticity and processing memory.3 Histone acetylation in insular cortex causes formation of memories. HDAC inhibitors are reported to increases the magnitude of CA1 synapses. It is therefore hypothesized that memory enhancement might be possible by HDAC inhibitions. In a study it has been seen that in vitro HDAC inhibitor administration or increase in acetylation at H3K9 or H3K14 and methylation in H3K4, both the cases could reinstate the lost memory after brain atrophy and neuronal loss.4
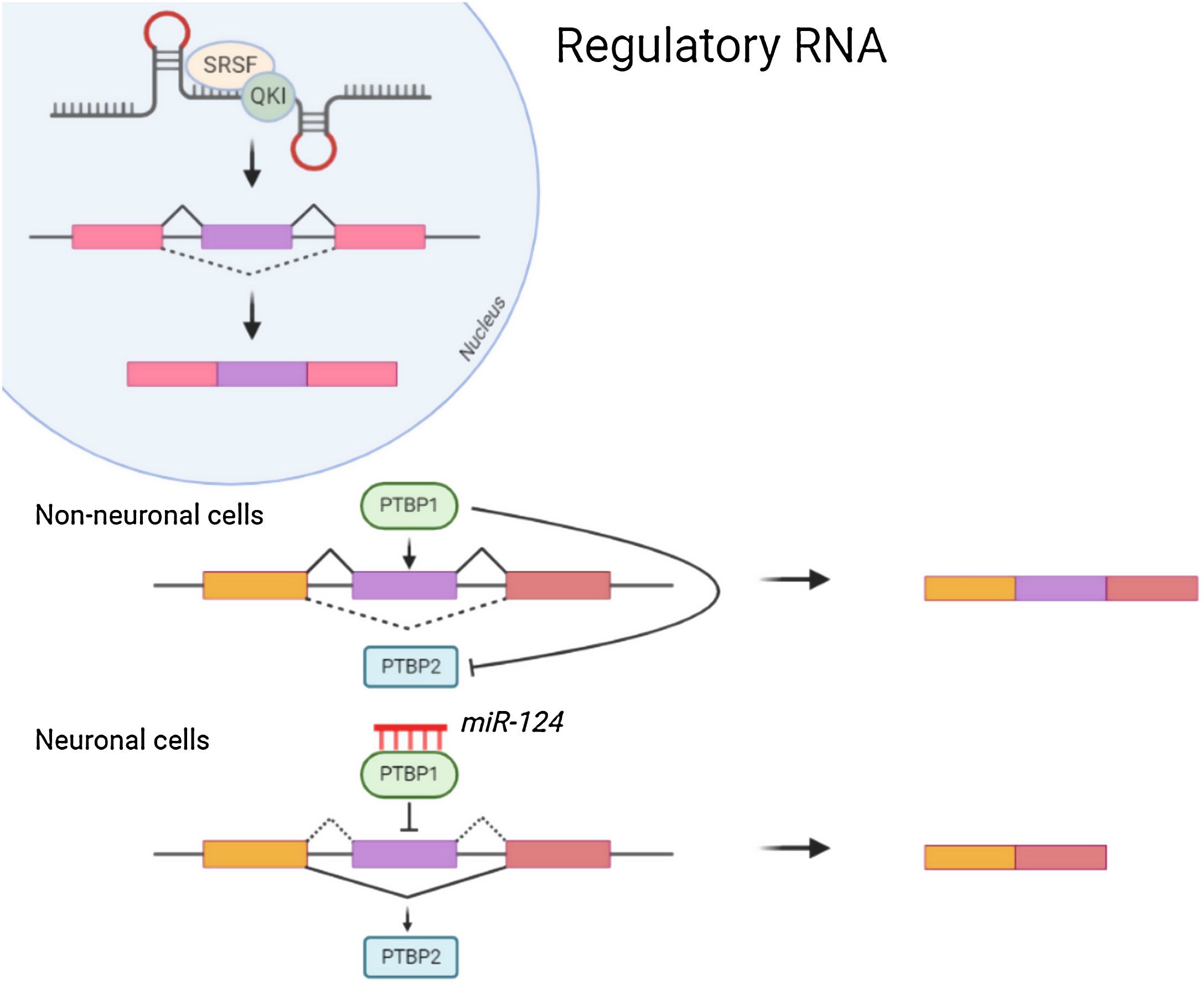
Non-coding RNANon-coding RNA such as MicroRNAs(miRNA) attach to the 3′ Untranslated region (UTR) of mRNAs for mediating post transcriptional regulation (inhibiting the translation or degradation). The miRNA-124 contributes towards maturation and differentiation of neuronal cells. Repression of PTBP1 by miRNA-124 (Fig. 1) leads to the formation of neuronal cells which instead might form non neuronal cells. Also, Some of the non-coding RNA are reported to be essential for dendritogenesis, synapse formation and maturation.5 Neurone specific miRNA also regulates the local protein synthesis needed for neuronal survival and synaptic plasticity. Long non coding RNAs enhances the synapse related gene expression in hippocampal neurons.6
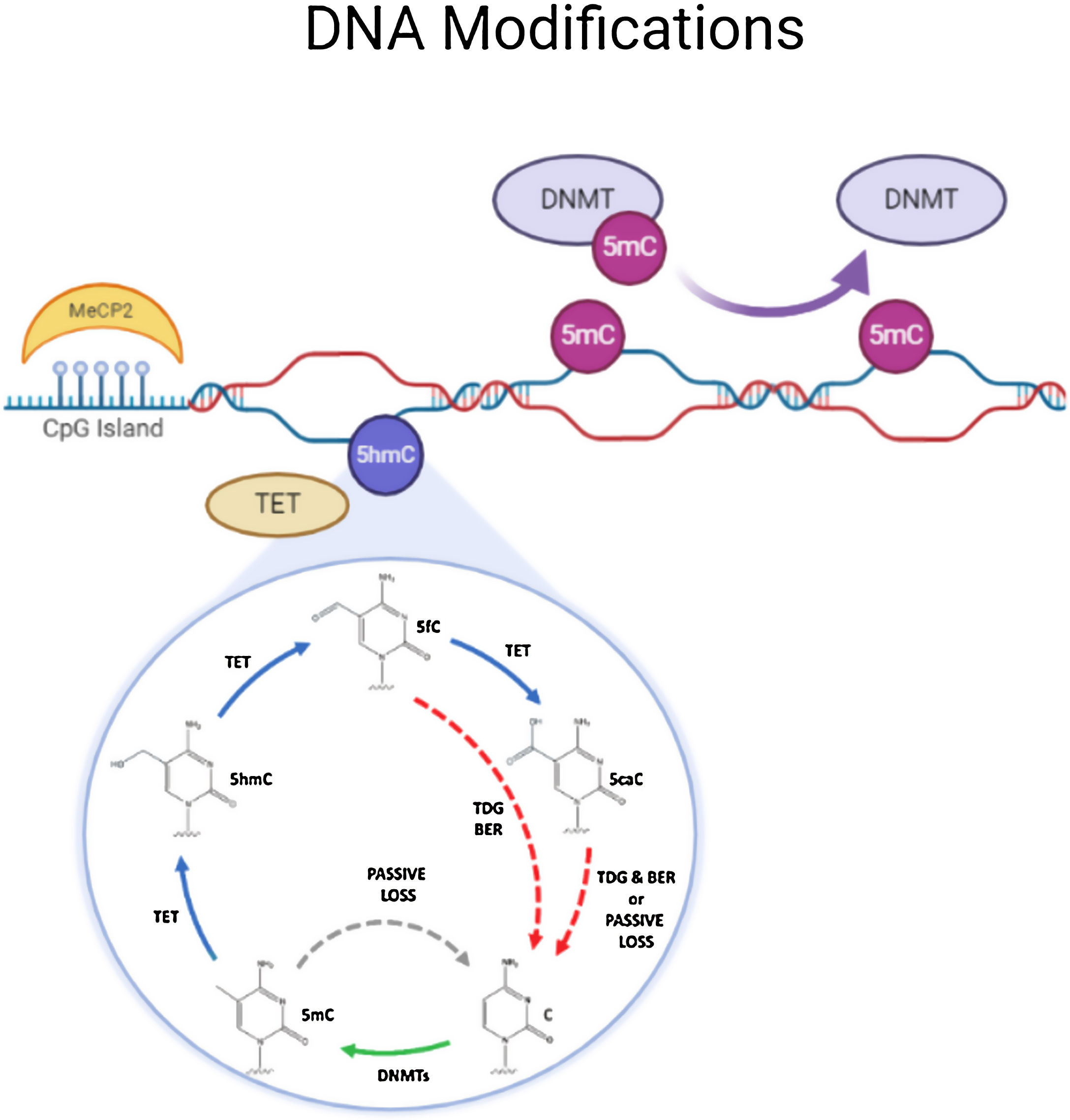
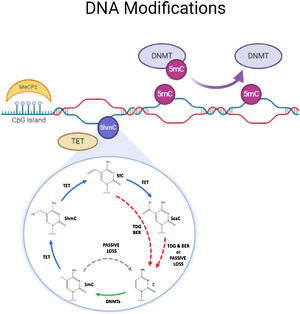
DNA methylationDNA methylation is responsible of the silencing of gene expression. It has been seen that DNA methylation is crucial for memory acquisition and storage.7 Methylation occurs due to inheritance as well as in response to environmental factors, normal aging and diseases. Not only at cytosine-phosphate-guanine (CpG) island but it has also been seen that DNA methylation also occur in non-CpG (CpH) sites. This CpG methylation are seen is huge number in the neuronal gene because of this restraining transcription. DNA methyl transferases (DNMT) such as DNMT1, DNMT3A and DNMT3B cause methylation on DNA. In one of the study it has been shown that when DNMT1 and DNMT3A are deleted in a mice, they found loss of hippocampus-based learning and memory.8 Conversion of 5-methyl-cytosine (5mC) to 5-hydroxymethylcytosine (5hmC) in synaptic plasticity and memory associated genes are brought by methylcytosine dioxygenases ten-eleven translocation proteins (TET).9 TET is responsible to covert 5mC into 5-formyl-cytosine(5fC), 5hmC and 5-carboxy-cytosine (5caC) (Fig. 2). Hence, these genes cause DNA demethylation and activate the genes responsible for memory.
Epigenetics and neurodegenerationMajority of the neurodegenerative disorders occur during aging. Therefore, over time various factors affect the epigenetic modifications for example cigarette smoking, food habits, exercise etc. These epigenetic changes might cause death of neurons thus leading to various neurodegenerative disorders. The modifications in neurons are not passed to new generation.
Parkinson's diseasePD is caused by the death of dopaminergic neurons containing neuromelanin in the substantia nigra in the brain, with the presence of Lewy bodies. PD results from combination of polygenic inheritance, environmental exposure and also complex gene-environment interaction.10 PARK, α-synuclein (SNCA), DJ-1, Parkin, leucine-Rich receptor kinase 2 are the genes responsible for PD.
When epigenetic analysis has been done on PD brain it revealed that SNCA gene might be regulated epigenetically.11 Hypermethylation in SNGA has been found in individuals with alcoholism and anorexia12 hence giving evidence of epigenetic regulation of SNCA. Reduction of DNA methylation in intron 1 of SNCA was observed in several regions of brain of sporadic PD patient.13 Increased level of SNCA expression is seen if DNA methylation is inhibited. It was recently found out that sequestration of DNMT1 from the nucleus is done by α-synuclein.14 As we know DNMT1 is responsible for demethylation thus increasing the concentration of α-synuclein in the brain.
Unlike SNCA, promoter methylation of Parkin gene does not cause any effect in development or pathogenesis of PD.15
Histone modificationsStudies showed that inhibition of histone acetylation happens when α-synuclein binds to histone16 therefore HDAC inhibitors protects the neurone against neurotoxicity mediated by α-synuclein. It was found that in Drosophila α-synuclein binds to H3 and causes inhibition of histone acetylation.17 In numerous PD models decrease in α-synuclein mediated neurotoxicity were observed by inhibition of histone deacetylase Sirtuin 2.18 In human α-synuclein overexpression can downregulate the nine genes forming histones H1, H2B and H4 had been reported.19 Therefore, we can conclude that histone modification has crucial role to play in PD.
Epigenetic changes by non-coding RNAsThe regulation of maturation and function of dopaminergic neurons in midbrain is done by the microRNA miR-133b. In the midbrain of PD patient's deficiency of miR-133b was seen.20 miR-7 and miR-153 together regulates the concentration of α-synuclein. Both miRNAs bind to 3’UTR of α-synuclein and downregulates both transcription and translation.21 In C. elegans PD model underexpression of several miRNA was analysed.22 So overall studies revelled miRNA also regulates PD pathogenesis.
DNA methylationPyrosequencing of the brain region showed differences in levels of DNA methylation in substantia nigra in CpG islands.23 The bisulfite sequencing is the most recent and “gold standard” method to find out DNA methylation. It has been noticed that environmental and nutritional factors like manganese(Mn), coffee, pesticides, deficiency of folate and some single nucleotide polymorphism plays vital role in DNA methylation.24 A study using SHAY5Y cells(dopaminergic human neuroblastoma) where, alteration in function of PARK2 and PINK1 due to hypermethylation of DNA were observed upon exposure to Mn.25 Hypermethylation in SNCA, NPAS2, PGC1-α and CYP2E1 genes are also responsible for PD.
Alzheimer's diseasesAD is most common among mid 1960s. It is a neurodegenerative disorder which slowly destroys the memory and thinking skill. Among the aged people it causes dementia. Accumulation of amyloid-β(Aβ) plaques and neurofibrillary tangles are the important characteristics of AD. Amyloid precursor protein (APP), presenilin 1 or 2 genes if get mutated causes autosomal dominant AD.
Histone modificationsClass 1 HDACs (HDAC2 & HDAC3) are expressed in higher concentration in region of brain associated with memory. Enhancement of microglial amyloid phagocytosis was observed in an AD mouse model when class 1 HDACs genes are deleted.26 HDAC2 decreases the histone acetylation of genes responsible for memory and learning.27 In AD mouse model overexpression of HDAC2 in prefrontal cortex and hippocampus was reported in a study.28 52% elevation of HDAC6 in cortex and 91% in hippocampus were found in AD patients29 causing cognitive impairment. These studies indicate that histone modification play a vital role in AD development.
Epigenetic changes by non-coding RNAsNon coding RNAs show a huge difference in their level of expression in healthy and AD brain. 555 circular RNAs, 183 miRNAs and 319 mRNAs in an AD rat model were found to be significantly downregulated.30 In AD pathogenesis miRNAs helps in amyloid processing, insulin resistance and neurogenesis. In a human AD brain, it was seen that both synaptic plasticity and memory could be regulated by brain derived neuropathic factor which in turn tuned by miR-206.31
DNA methylationMethylation of 5mC in DNA has important role in neuronal gene expression and neuronal development. These products play critical role development and function of neurons. In AD patients it has been seen that signatures of 5mC, 5Fc, 5hmC and 5caC are limited32 hence causes degeneration of neurons. Cytosines at 207-182 region of APP are methylated in normal brain but with aging this cytosines get demethylated and led to deposition of Aβ in the brain. It was found that in hippocampus, cerebellum and entorhinal cortex of AD patient levels of 5mC were reduced.33
Huntington's diseaseHD is a neurodegenerative genetic disease which affects medium spiny nerves of striatum. It is caused by unsteady expanded CAG repeats(more than 35 repeats) in Huntingtin gene (HTT), resulting in the synthesis of mutant protein (mHtt) with a tract of toxic polyglutamine (polyQ).34 Longer the expansion of CAG more sever affects are seen in HD patients. In long run neuronal dysfunction caused by HD proceeds to death of neurons. Depression, mania, suicide, apathy, dystonia, motor skill learning deficits are the consequences of HD.
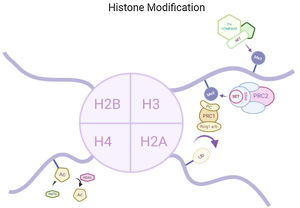
Histone modificationsHtt overexpression led to extensive changes in level of histone acetylation. In the striatum of HD mice decrease in acetylation of H3 is seen at the promoter of Drd2, Penk1, Actb or Grin1 genes which are normally underexpressed for the function of neurons present in striatum. These genes are upregulated due to the acetylation.35 In some studies, increase in level of H3K9me2 and H3K9me3 were found which causes heterochromatin structure in the striatum in HD patient.36,37 In the frontal cortex and blood of patients suffering from HD it has been that a member Y of H2A histone family(H2AFY) is overexpressed.38 Increase in monoubiquitylation at H2A (uH2A) is seen in striatum of HD patient which has the ability to influence transcription (Fig. 3). Hypoacetylation in H3K9,14 and H4K12 has also been reporter in mice model.39
DNA methylationAt both promoter proximal and distal regulated regions modification in DNA methylation was reported due to the mutant Htt.40 Further studies gave the idea of modified level of DNA methylation in HD brain tissue. Although many studies have been done so far, there are no significant amount of evidence suggesting a role of DNA methylation in HD.
Prion diseaseCreutzfeldt–Jakob Disease (CJD) is a prion disease prevalent in human which is close to bovine spongiform encephalopathy in cattle. CJD is one of the fatal neurodegenerative disorder having expeditious progression. This disease prior to its onset leads to death of the patients within a year. A specific conformational change in Prion protein (PrP) or misfolded PrP results in infectious prion protein. It causes spongiform degeneration of grey matter besides proliferation of astrocytes. Fatal familial insomnia is another sporadically occurring inherited prion disease in human. As the disease is associated with protein folding rather than any nucleic acid changes it reveals unusual form of epigenetics. Abrupt change in PH, temperature and intracellular metabolites causing environmental stress. The frequency of prion switching increases with the extreme stresses, subsequently epigenetic changes occurring in extreme conditions.41
ConclusionThese studies gives us an idea how the epigenetic changes impact the neuronal cells and the diseases associated with them. Throughout many years researchers mainly focused on the genomic changes associated with the diseases but neuroepigenetic related modifications were mostly neglected. Novel therapeutic treatment for the neurodegenerative diseases can be done by reversing the epigenetic changes. In most of the cases it has been seen that HDAC plays a major role in the memory loss as well as in neurodegenerative disorders. On the ground of this, drug inducing HDAC inhibitor might result in epigenetic reversion. In an experimental study it was shown that inhibitor of Class IIa HDACs provides a promising therapeutic strategy for treating HD.42 In many studies the role of HDAC3 in AD progression has been shown therefore inhibitor of HDAC3 is having great potential in therapeutic treatment of AD. RGFP-966 is an inhibitor of HDAC3 was found to be associated with huge number of activities in both improving memory and AD reversion.43 Although in a rat model of PD upon administration of nicotinamide used for inhibition of HDAC class III many adverse effects were observed such as upregulation of neurotrophic and neuroprotective factors as well as histone hyperacetylation. Thereupon, the inhibitor of HDAC instead of neuroprotection exhibited neurodegeneration.44 Another epigenetic modification includes DNA methylation where hypermethylation in CpG island is seen in the neuronal cells. Drugs targeting DNA methyltransferases and effectivity were reported, suggesting novel target for therapeutic treatment of neurodegenerative disorders. Decitabine and FdCyd were reported as efficient. inhibitor of DNA methyltransferase in HD mouse model suggesting reduction in neurotoxicity.45 Although many studies focused on the reversion of epigenetic modifications but some of these lacked targeted drug delivery thus resulting in promotion of degeneration of neuronal cells. More focus on target specific drug delivery should be done in future.
Conflict of interestsThere is no conflict of interests.