The expression of specific miRNAs and their mRNA targets are changed in infectious disease. The aim of this study was to analyze the expression of pro-neuroinflammatory miRNAs, anti-neuroinflammatory miRNAs, and their mRNA targets in the serum of COVID-19 patients with different grades.
MethodsCOVID-19 patients with different grades were enrolled in this study and the expression of pro-neuroinflammatory miRNAs, anti-neuroinflammatory miRNAs, and their target mRNAs was analyzed by q-PCR.
ResultsThe relative expression of anti- neuroinflammatory miRNAs (mir-21, mir-124, and mir-146a) was decreased and the relative expression of their target mRNAs (IL-12p53, Stat3, and TRAF6) was increased. Also, the relative expression of pro-neuroinflammatory miRNAs (mir-326, mir-155, and mir-27b) was increased and the relative expression of their target mRNA (PPARS, SOCS1, and CEBPA) was decreased in COVID-19 patients with increase of disease grade. A negative significant correlation was seen between mir-21 and IL-12p53 mRNA, mir-124 and Stat3 mRNA, mir-146a and TRAF6 mRNA, mir-27b and PPARS mRNA, mir-155 and SOCS1 mRNA, and between mir-326 and CEBPA mRNA in COVID-19 patients (P<0.05).
ConclusionsThis study showed that the relative expression of anti- neuroinflammatory miRNAs was decreased and the relative expression of their targeted mRNAs was increased in COVID-19 patients from asymptomatic to critical illness. Also, this study showed that the relative expression of pro-neuroinflammatory miRNAs was increased and the relative expression of their targeted mRNA was decreased in COVID-19 patients from asymptomatic to critical illness.
La expresión de miARN específicos y sus dianas de ARNm se modifican en las enfermedades infecciosas. El objetivo de este estudio fue analizar la expresión de miARN pro-neuroinflamatorios, miARN anti-neuroinflamatorios y sus ARNm dianas en el suero de pacientes con COVID-19 de diferentes grados.
MétodosSe incluyeron en este estudio pacientes con COVID-19 de diferentes grados y se analizó la expresión de miARN pro-neuroinflamatorios, miARN anti-neuroinflamatorios y sus ARNm diana mediante q-PCR.
ResultadosLa expresión relativa de miARN anti-neuroinflamatorios (mir-21, mir-124 y mir-146a) disminuyó y la expresión relativa de sus ARNm diana (IL-12p53, Stat3 y TRAF6) aumentó. Además, la expresión relativa de miARN pro-neuroinflamatorios (mir-326, mir-155 y mir-27b) aumentó y la expresión relativa de su ARNm diana (PPARS, SOCS1 y CEBPA) disminuyó en pacientes con COVID-19 con aumento del grado de enfermedad. Se observó una correlación negativa significativa entre ARNm de mir-21 e IL-12p53, ARNm de mir-124 y Stat3, ARNm de mir-146a y TRAF6, ARNm de mir-27b y PPARS, ARNm de mir-155 y SOCS1, y entre mir-326 y ARNm de CEBPA en pacientes con COVID-19 (p < 0,05).
ConclusionesEste estudio mostró que la expresión relativa de miARN anti-neuroinflamatorios disminuyó y la expresión relativa de sus ARNm diana se incrementó en pacientes con COVID-19 de enfermedad asintomática a crítica. Además, este estudio mostró que la expresión relativa de miARN pro-neuroinflamatorios aumentó y la expresión relativa de su ARNm diana disminuyó en pacientes con COVID-19 de enfermedad asintomática a crítica.
Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2), known as COVID-19, is a new infectious disease first seen in late December 2019 in Wuhan, China, and similar outbreaks occurred in the hospital in neighboring countries. Major clinical symptoms include fever, dry cough, diarrhea, muscle aches, pneumonia, and in severe cases death.1,2 COVID-19 also is associated with neurological manifestations such as encephalopathy and encephalomyelitis, ischemic stroke and intracerebral hemorrhage, anosmia, neuromuscular diseases, and neuroinflammation diseases.3
Since COVID-19 is a new disease, complete information on its etiology, cellular mechanisms, and possible risk factors is not available. COVID-19 may be similar to recent acute respiratory syndromes, such as SARS and MERS.4 Theoretically, after the SARS-CoV-2 enters the human body, different types of immune cells are stimulated. These cells trigger the proper immune response by producing different cytokines, chemokines, antibodies, etc. SARS-CoV-2 can infect the CNS following the entry of the virus into the nose or the eye. The viral particles are transmitted to the olfactory bulb and then to the brainstem, and then all parts of the brain.5 In addition to the direct attack of nerve cells, the SARS-CoV-2 can systematically cross the BBB through the blood vessels and reach the CNS. The main feature of systemic infection in COVID-19 is the massive increase in pro-inflammatory factors in the blood, which is described as a “cytokine stor”.6 This leads to BBB permeability and transmission of SARS-CoV-2 and peripheral immune cells. Once the coronavirus enters the CNS, it is the turn of the astrocytes and microglia to fight it. The immune response of astrocytes and microglia is regulated by different microRNAs (miRNAs). Previous studies showed inflammatory processes in CNS are guided by pro-neuroinflammatory miRNAs (such as mir-155, mir-27b, mir-326) and anti- neuroinflammatory miRNAs (such as mir-146a, mir-124, and mir-21).7,8
This study aimed to analyze the expression of pro-neuroinflammatory miRNAs, anti-neuroinflammatory miRNAs, and their mRNA targets in the serum of COVID-19 patients with different grades.
Materials and methodsMaterialsAll primers were provided from Bioneer, South Korea. MirPremier microRNA isolation kit was sourced from Sigma-Aldrich, USA. Mir-X miRNA First-Strand Synthesis kit and cDNA matermix were purchased from Takara bio inc, USA. SYBR® Green Real-Time Master Mix was from Invitrogen, UK.
BioinformaticsIn this study, to determine the miRNAs associated with the COVID-19, we used online bioinformatics Softwares.9 In the first step, mirTarP (https://mcube.nju.edu.cn/jwang/mirTar/docs/mirTar/) was used to the list of appropriate miRNAs.10,11 In the second step, to reduce the number of selected miRNAs, we selected some limited pro-neuroinflammatory and anti-neuroinflammatory miRNAs that were previously reported in other studies. In the third step, the miRDB online database (http://mirdb.org/) was used to find the target of selected miRNAs.12 Target genes of the differentially regulated miRNAs were predicted using the mirPath tool (version 3.0).13 KEGG molecular pathways were also retrieved using the same tool.14 Pathways and processes regulated with P values lower than 0.05 were considered significant.
Study groupsTable 1 shows the full characteristics of 6 study groups enrolled in this study. The licensing committee that approved the experiments, including any relevant details was Zahedan University of Medical Sciences, Zahedan, Iran. All experiments were under the guidelines of the National Institute of Health, and the ethics committee of Zahedan University of Medical Sciences, Zahedan, Iran. (Ethical code: IR.ZAUMS.REC.1399.317). Also, informed consent was obtained from all participants. Five ml of whole blood was collected from each person and their serum was separated by centrifugation at 3000rpm/min for 10min at 4°C. In this study, only COVID-19 patients with English variant of SARS-COV-2 (Lineage B.1.1.7; GISAID accession number: EPI-ISL-2227268) were included.
The characteristics of study groups.
| Study group 1 | Study group 2 | Study group 3 | Study group 4 | Study group 5 | Control | |
|---|---|---|---|---|---|---|
| Number (n) | 21 | 20 | 20 | 21 | 21 | 20 |
| Age distribution±SD | 50±10 | 50±10 | 50±10 | 50±10 | 50±12 | 50±12 |
| Sex percentage±SD | Female (52%±2%)Male (48%±1%) | Female (51%±2%)Male (49%±2%) | Female (50%±3%)Male (50%±1%) | Female (53%±1%)Male (47%±2%) | Female (52%±1%)Male (48%±2%) | Female (50%±3%)Male (50%±1%) |
| Severity of illnessa | Grade 5Critical illness: Individuals who have respiratory failure, septic shock, and/or multiple organ dysfunction. | Grade 4Severe illness: Individuals who have SpO2<94% on room air at sea level, a ratio of arterial partial pressure of oxygen to fraction of inspired oxygen (PaO2/FiO2)<300mm Hg, respiratory frequency >30 breaths/min, or lung infiltrates >50%. | Grade 3Moderate illness: Individuals who show evidence of lower respiratory disease during clinical assessment or imaging and who have saturation of oxygen (SpO2) ≥94% on room air at sea level. | Grade 2Mild illness: Individuals who have any of the various signs and symptoms of COVID-19 (e.g., fever, cough, sore throat, malaise, headache, muscle pain, nausea, vomiting, diarrhea, loss of taste and smell) but who do not have shortness of breath, dyspnea, or abnormal chest imaging. | Grade 1Asymptomatic: Individuals who test positive for SARS-CoV-2 using a virologic test (i.e., a nucleic acid amplification test or an antigen test) but who have no symptoms that are consistent with COVID-19. | Healthy people |
| Comorbidities | No | No | No | No | No | No |
| Inflammatory autoimmune diseases | No | No | No | No | No | No |
| Drug treatment | No | No | No | No | No | No |
Here, Small RNA was isolated from blood samples using mirPremier microRNA isolation kit. Briefly, 1000μL of the lysis buffer was added to 100μL of serum samples, vortexed for 2min, and incubated at 55°C for 5min. The samples were then centrifuged for 5min at 14,000×g to remove cellular debris, genomic DNA, and large RNA. The lysate supernatant was filtered through the filtration column and binding column. After binding, the column was first washed with 700μL of 100% ethanol and centrifuged at 14,000×g for 30s and again the flow-through was discarded. The second wash was done by adding 500μl of binding solution into the column and centrifuged at maximum speed (14,000×g) for 1min. Subsequently, 500ml of the ethanol-diluted wash solution 2 was added to the column for a third wash. After centrifugation at maximum speed (14,000×g) for 30s, the flow-through was discarded. Next, the column was dried by centrifuging at maximum speed (14,000×g) for 1min. The column-tube assembly was carefully removed from the centrifuge to avoid splashing of the residual flow-through liquid to the dried column. Small RNA was eluted from the column using 50ml elution solution and by centrifugation at 16,000×g and the process was repeated to improve small RNA yield. The purity of the RNA samples was analyzed by NanoDrop ND-1000 UV-VIS spectrophotometer. The A260nm/A280nm ratio of all samples was between 1.8 and 2.1. The quantity of RNA samples was analyzed by agarose gel electrophoretic separation. For first-strand cDNA synthesis, small RNAs were polyadenylated and reverse transcribed using the Mir-X miRNA First-Strand Synthesis kit. Briefly, 5μl mRQ buffer (2×), 5μg RNA and 1.25μl mRQ enzyme was mixed in a reaction volume of 10μl and incubated in a thermocycler for 1h at 37°C, then terminate at 85°C for 5min to inactivate the enzymes. After reverse transcription, the cDNA was diluted. For quantification of miRNA by qPCR, Mir-X miRNA qPCR SYBR Kit was used. Briefly, 10μl PCR reaction mixture was prepared to comprise of 1× SYBR advantage premix, 0.2mM of both forward and reverse primers, and 50ng of the first-strand cDNA. qPCR reactions were incubated in a 96 well plate at 95°C for 2min, followed by 40 cycles of 95°C for 10s and 60°C for 20s. Amplification cycles were followed by a melting curve analysis ranging from 56 to 95°C. Finally, the threshold cycle (Ct) values were recorded. For mRNA, total RNA was extracted using an RNA extraction kit. Then, the cDNA was synthesized in the presence of the superscript enzyme and hexamers. For real-time PCR, 2μL of cDNA, 2μL of forward primer, and 2μL of reverse primer of each gene were added to 10μL of SYBR® Green Real-Time Master Mix. In this study, the relative expression of mir-155, mir-27b, mir-326, mir-124, mir-146a, mir-21, IL-12p53, Stat3, TRAF6, PPARS, SOCS1, and CEBPA was analyzed. The expression of microRNA and mRNA was normalized to RNU 48 and GAPDH, respectively.
Statistical analysisAll data were reported as the mean±standard deviation. To find significant differences between groups, a one-way ANOVA method was applied. A P-value of less than 0.05 was considered statistically significant. Also, Spearman's correlation coefficient was used to correlate the expression of miRNAs and their mRNA targets.
ResultsBioinformatics analysisFive-top human mRNA targets for pro-neuroinflammatory miRNAs (mir-155, mir-27b, and mir-326) and anti- neuroinflammatory miRNAs (mir-124, mir-146a, and mir-21) are shown in Table 2. It should be noted that each miRNA has many targets, but here we have listed only 5 important mRNA targets with the highest target score. Theoretically, all of them can be affected by pro-neuroinflammatory and anti-neuroinflammatory miRNAs.
The human gene targets of pro-neuroinflammatory miRNAs and anti- neuroinflammatory miRNAs, obtained from miRDB online database.
| Target score | miRNA Name | Gene Symbol | Gene description |
|---|---|---|---|
| 98 | miR-155 | SOCS1 | Suppressor Of Cytokine Signaling 1 |
| 99 | miR-155 | ZNF629 | Zinc finger protein 629 |
| 99 | miR-155 | CREBRF | CREB3 regulatory factor |
| 99 | miR-155 | DENND1B | DENN domain containing 1B |
| 98 | miR-155 | PTPN21 | Protein tyrosine phosphatase, non-receptor type 21 |
| 98 | miR-27b | PPARs | Peroxisome Proliferator Activated Receptor Gamma |
| 97 | miR-27b | AFF4 | AF4/FMR2 family member 4 |
| 97 | miR-27b | GXYLT1 | Glucoside xylosyltransferase 1 |
| 97 | miR-27b | ARFGEF1 | ADP ribosylation factor guanine nucleotide exchange factor 1 |
| 96 | miR-27b | GCC2 | GRIP and coiled-coil domain containing 2 |
| 99 | miR-326 | CEBPA | CCAAT Enhancer Binding Protein Alpha |
| 99 | miR-326 | ETS1 | ETS proto-oncogene 1, transcription factor |
| 99 | miR-326 | CEP85 | Centrosomal protein 85 |
| 98 | miR-326 | FGF11 | Fibroblast growth factor 11 |
| 98 | miR-326 | GPD2 | Glycerol-3-phosphate dehydrogenase 2 |
| 98 | miR-124 | Stat3 | Signal Transducer And Activator Of Transcription 3 |
| 98 | miR-124 | OSBPL3 | Oxysterol binding protein like 3 |
| 98 | miR-124 | SLC50A1 | Solute carrier family 50 member 1 |
| 98 | miR-124 | ITGB1 | Integrin subunit beta 1 |
| 98 | miR-124 | SIX4 | SIX homeobox 4 |
| 100 | miR-146a | TRAF6 | TNF Receptor Associated Factor 6 |
| 100 | miR-146a | FOXC1 | Forkhead box C1 |
| 100 | miR-146a | CPLX2 | Complexin 2 |
| 100 | miR-146a | STXBP6 | Syntaxin binding protein 6 |
| 100 | miR-146a | ZFX | Zinc finger protein X-linked |
| 99 | miR-21 | IL-12p53 | Interleukin 12 p53 protein |
| 99 | miR-21 | STK38L | Serine/threonine kinase 38 like |
| 99 | miR-21 | PCDH19 | Protocadherin 19 |
| 99 | miR-21 | LAMP1 | Lysosomal associated membrane protein 1 |
| 99 | miR-21 | GRIA2 | Glutamate ionotropic receptor AMPA type subunit 2 |
Based on KEGG database (Table 3), we found that both pro-neuroinflammatory miRNAs and anti-neuroinflammatory miRNAs are significantly enriched in important cellular pathways, such as PI3K-Akt signaling pathway, mRNA surveillance pathway, mTOR signaling pathway, MAPK signaling pathway, Wnt signaling pathway, and AMPK signaling pathway.
Important pathways of pro-neuroinflammatory miRNAs and anti- neuroinflammatory miRNAs, extracted from KEGG molecular pathway.
| KEGG pathway | P-value | Genes | miRNAs |
|---|---|---|---|
| Adherens junction | 0.0001 | 30 | 6 |
| Endometrial cancer | 0.0001 | 24 | 6 |
| Small cell lung cancer | 0.0001 | 36 | 6 |
| Regulation of actin cytoskeleton | 0.0001 | 67 | 56 |
| Bladder cancer | 0.0002 | 21 | 4 |
| PI3K-Akt signaling pathway | 0.0002 | 105 | 5 |
| Shigellosis | 0.0002 | 26 | 4 |
| Thyroid hormone signaling pathway | 0.0003 | 44 | 4 |
| Lysine degradation | 0.001 | 15 | 3 |
| Non-small cell lung cancer | 0.001 | 23 | 4 |
| mRNA surveillance pathway | 0.001 | 34 | 4 |
| mTOR signaling pathway | 0.002 | 25 | 4 |
| Oocyte meiosis | 0.003 | 38 | 4 |
| Prolactin signaling pathway | 0.003 | 28 | 4 |
| Melanoma | 0.003 | 25 | 5 |
| Ubiquitin mediated proteolysis | 0.004 | 48 | 5 |
| Fatty acid metabolism | 0.004 | 11 | 3 |
| Fatty acid elongation | 0.004 | 6 | 3 |
| Arrhythmogenic right ventricular | 0.006 | 16 | 3 |
| Estrogen signaling pathway | 0.006 | 31 | 5 |
| Signaling pathways regulating of stem cells | 0.007 | 42 | 5 |
| Gap junction | 0.008 | 27 | 5 |
| Sphingolipid signaling pathway | 0.009 | 38 | 5 |
| Amoebiasis | 0.01 | 30 | 4 |
| Pantothenate and CoA biosynthesis | 0.01 | 6 | 3 |
| MAPK signaling pathway | 0.01 | 72 | 5 |
| Insulin signaling pathway | 0.02 | 45 | 5 |
| Wnt signaling pathway | 0.02 | 41 | 4 |
| Axon guidance | 0.02 | 37 | 4 |
| Pathogenic Escherichia coli infection | 0.02 | 21 | 5 |
| AMPK signaling pathway | 0.02 | 41 | 5 |
| Hepatitis C | 0.03 | 20 | 5 |
| Vibrio cholerae infection | 0.03 | 23 | 4 |
| Epithelial cell signaling in Helicobacter pylori infection | 0.04 | 54 | 4 |
| Platelet activation | 0.04 | 39 | 5 |
| Steroid biosynthesis | 0.04 | 5 | 3 |
| Salmonella infection | 0.04 | 27 | 5 |
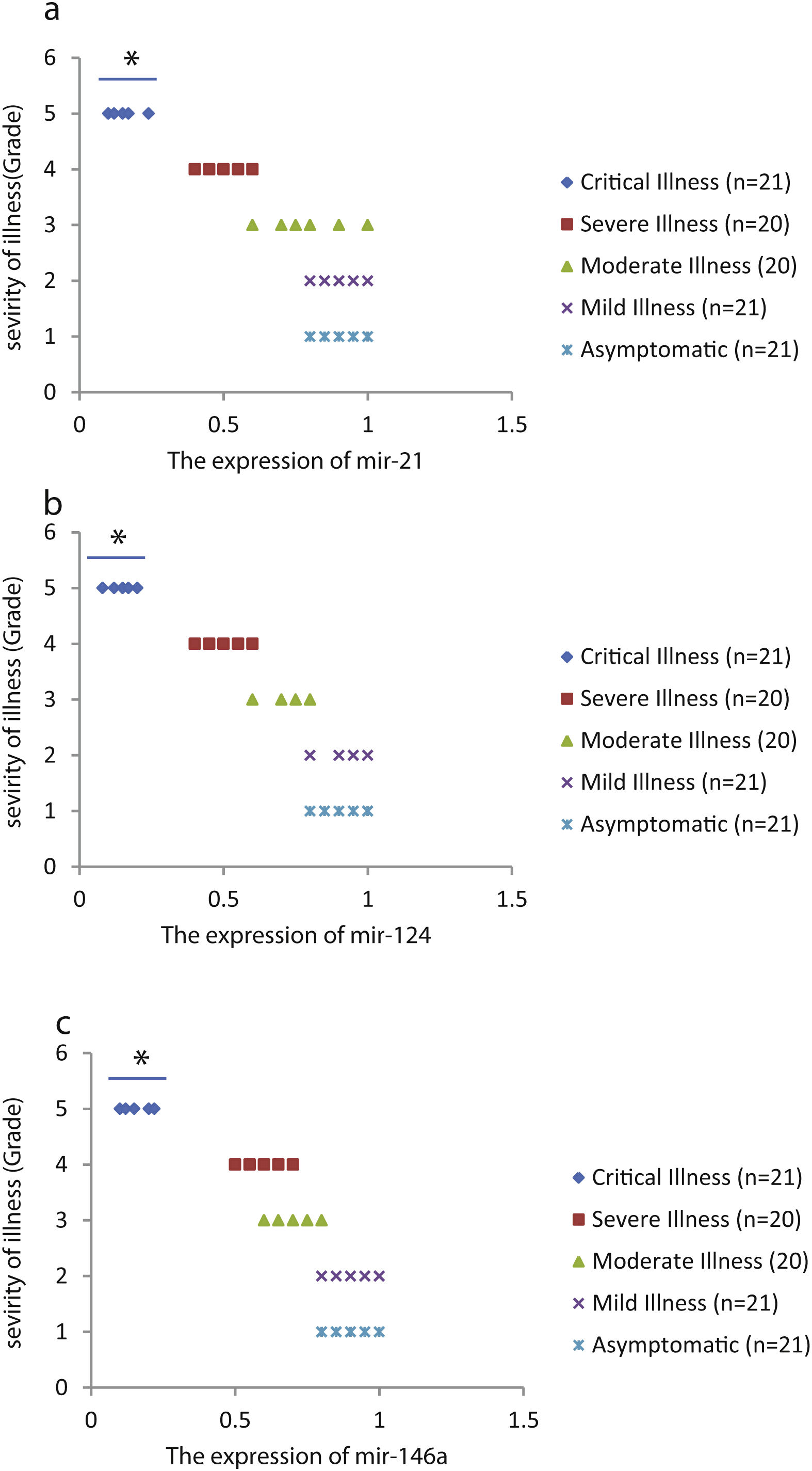
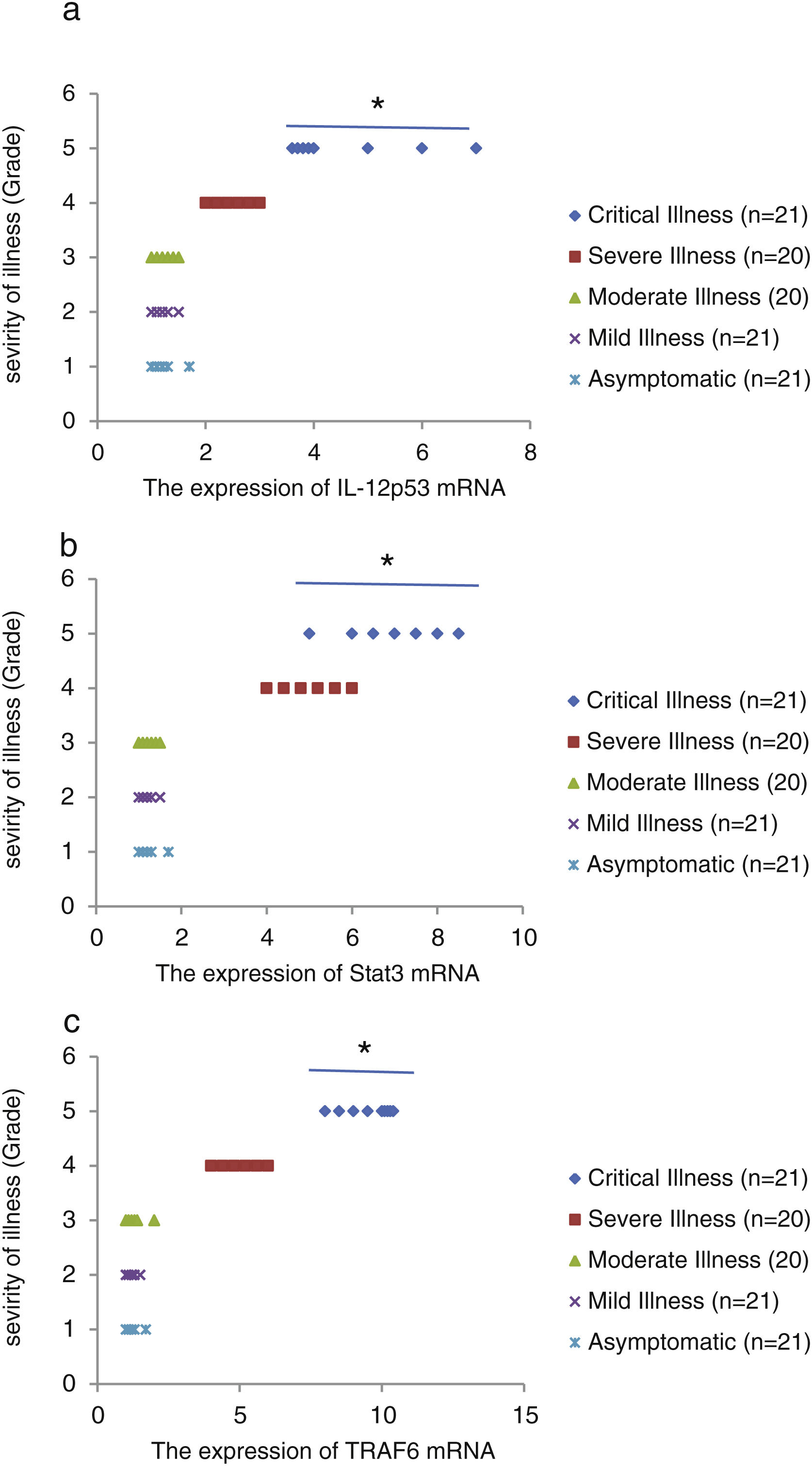
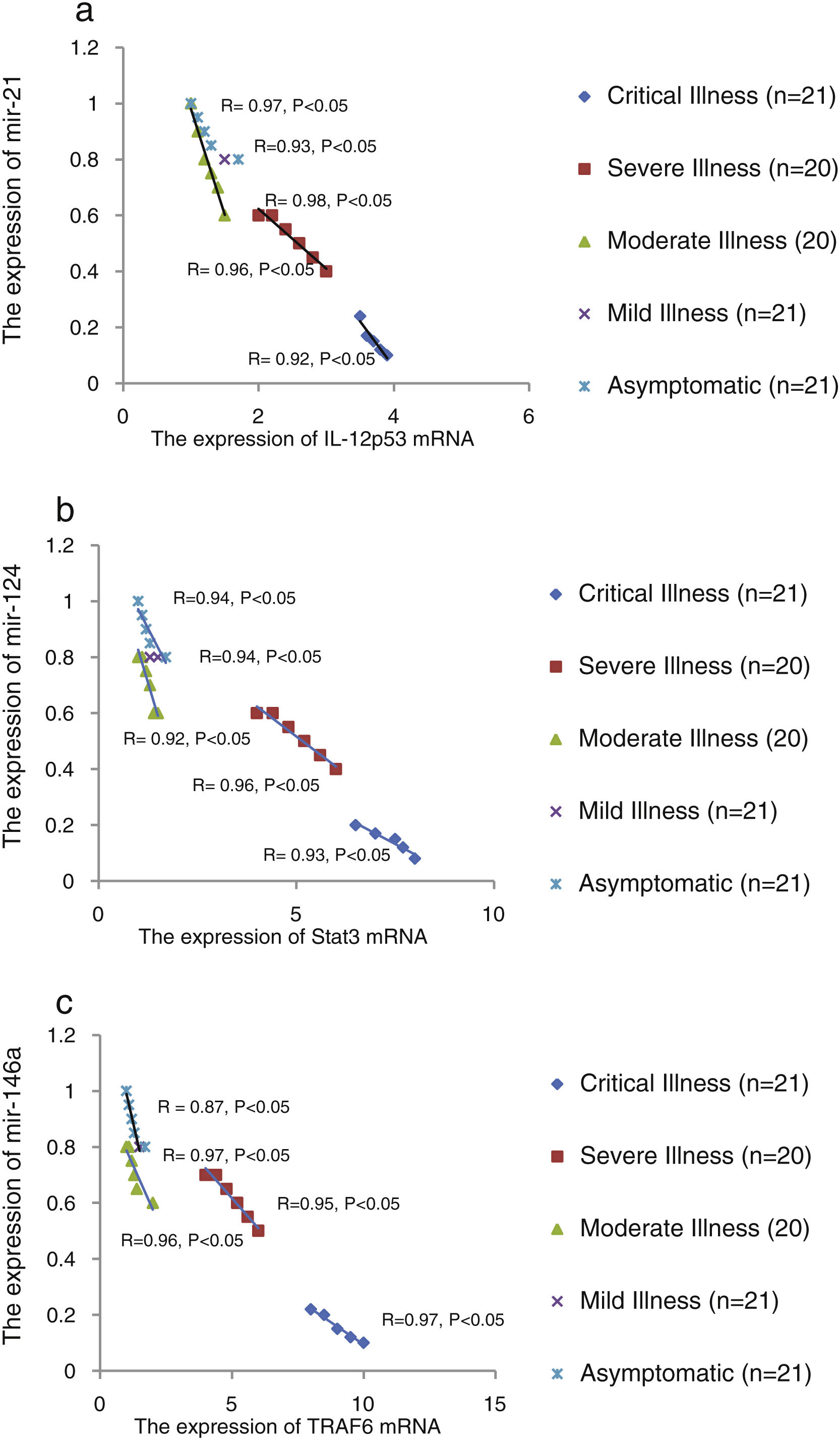
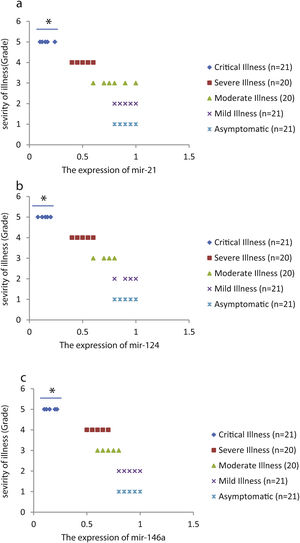
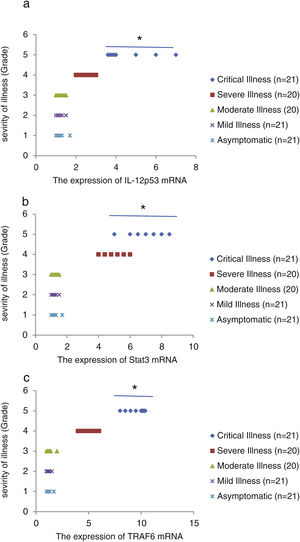
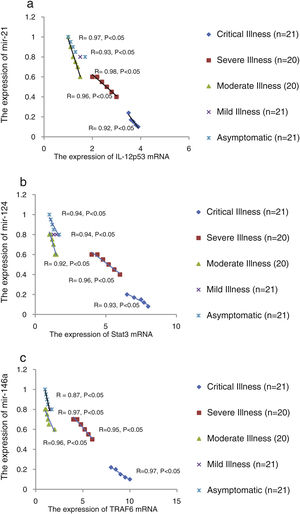
We found that the relative expression of anti-neuroinflammatory miRNAs, including mir-21, mir-124, and mir-146a, was significantly decreased with increase of COVID-19 grade (P<0.05) (Fig. 1(a–c)). Interestingly, the relative expression of human mRNA targets, including IL-12p53, Stat3, and TRAF6, of anti-neuroinflammatory miRNAs was significantly increased with increase of COVID-19 grade (P<0.05) (Fig. 2(a–c)). A negative significant correlation was seen between the expression of (mir-21 and IL-12p53 mRNA), (mir-124 and Stat3 mRNA), and (mir-146a and TRAF6 mRNA) in COVID-19 patients at all grades (P<0.05) (Fig. 3(a–c)).
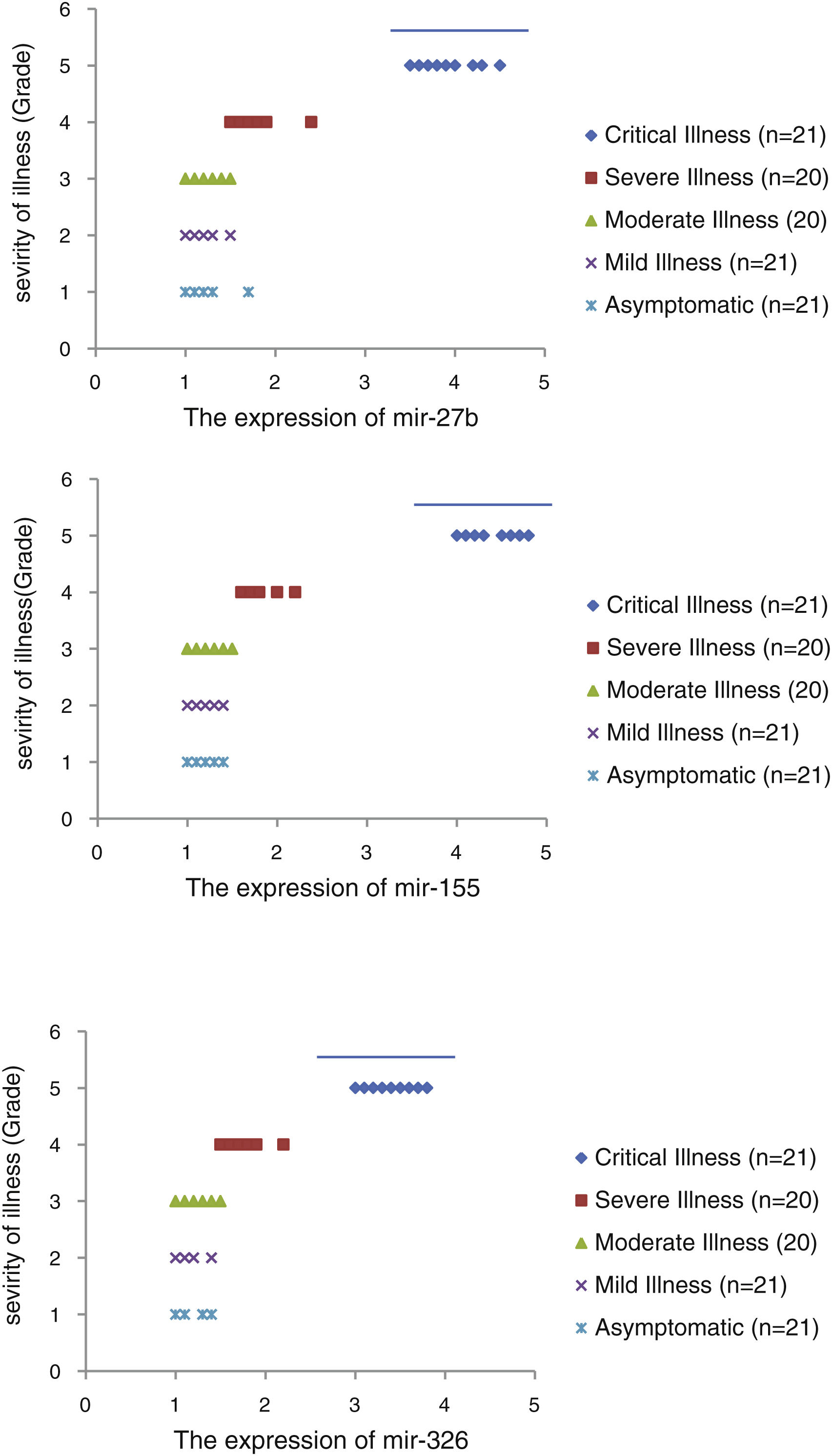
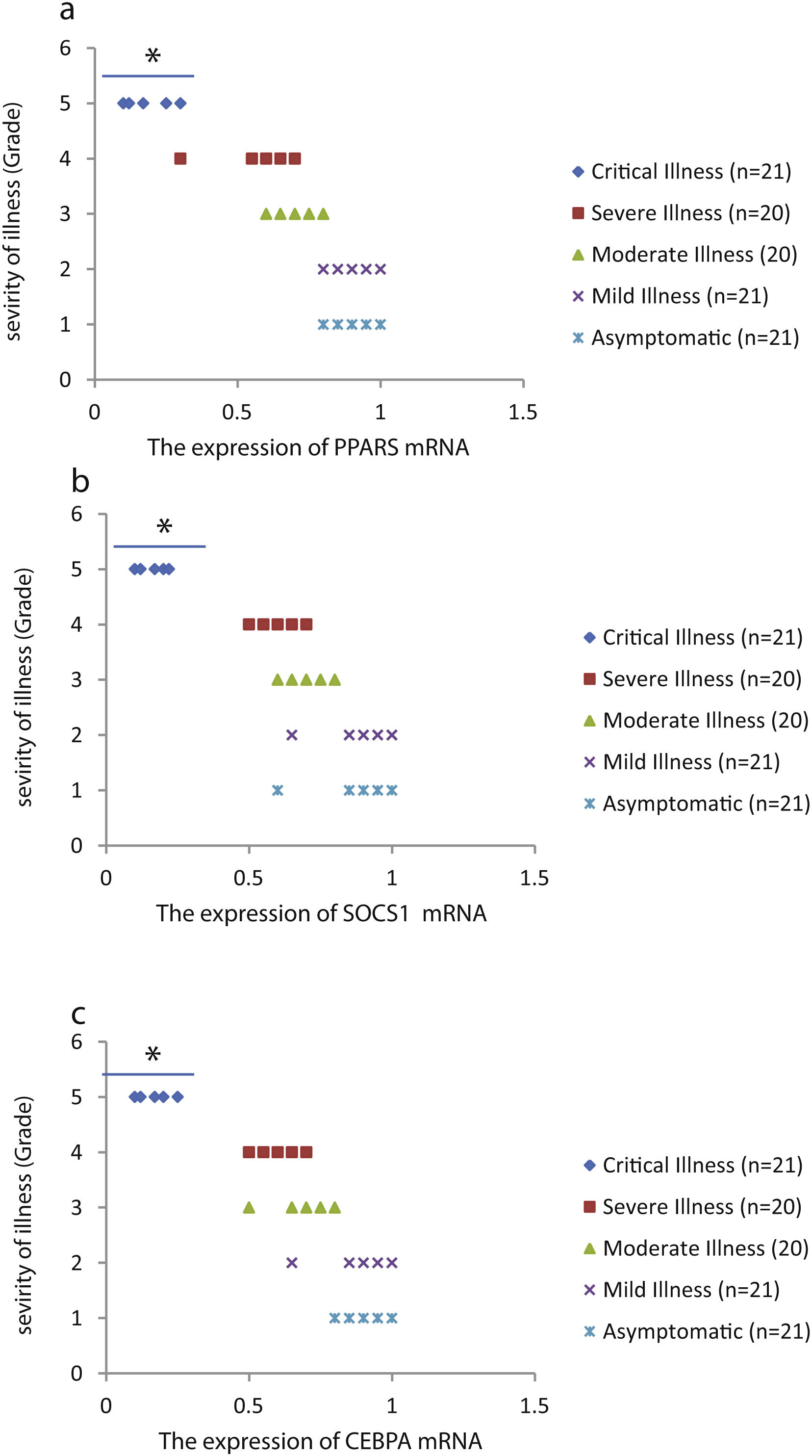
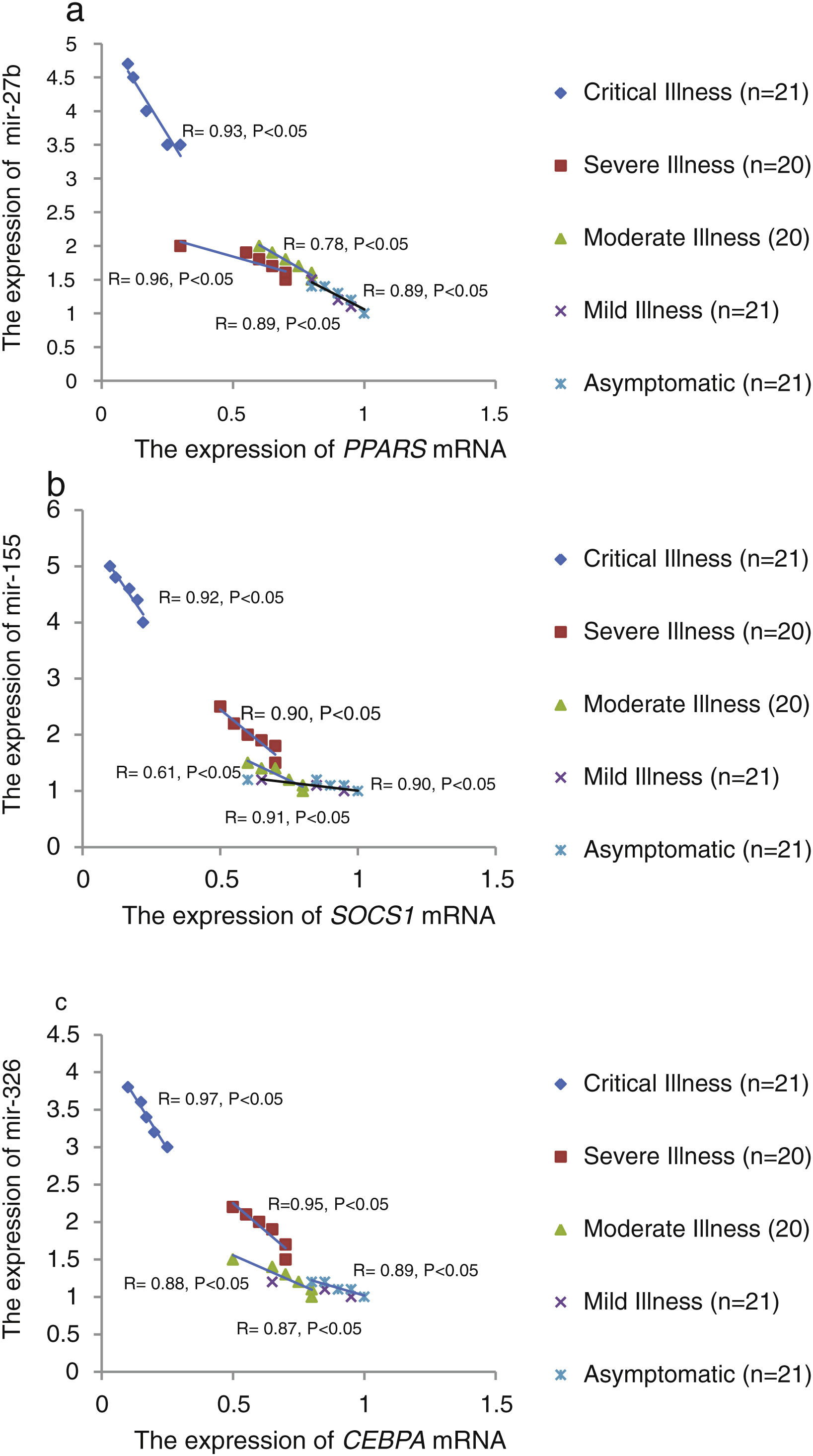
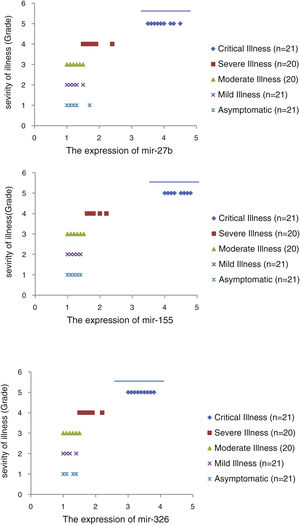
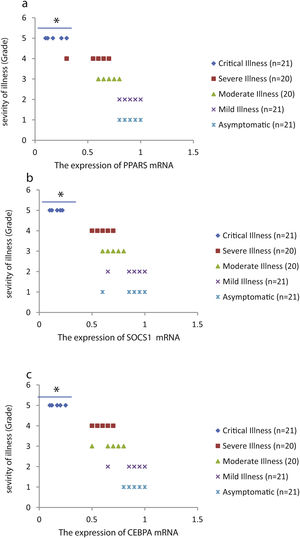
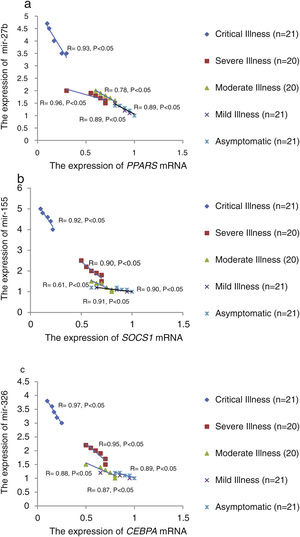
The relative expression of pro-neuroinflammatory miRNAs, including mir-326, mir-155, and mir-27b, was significantly increased with increase of COVID-19 grade (P<0.05) (Fig. 4(a–c)). Interestingly, the relative expression of human mRNA targets, including PPARS, SOCS1, and CEBPA, of pro-neuroinflammatory miRNAs was significantly decreased with increase of COVID-19 grade (P<0.05) (Fig. 5(a–c)). A negative significant correlation was also seen between the expression of (mir-27b and PPARS mRNA), (mir-155 and SOCS1 mRNA), and (mir-326 and CEBPA mRNA) in COVID-19 patients at all grades (P<0.05) (Fig. 6(a–c)).
This study showed that the relative expression of anti-neuroinflammatory miRNAs (mir-21, mir-124, and mir-146a) was decreased and the relative expression of their target mRNAs (IL-12p53, Stat3, and TRAF6) was increased in COVID-19 patients with increase of disease grade from asymptomatic to critical illness. Also, this study showed that the relative expression of pro-neuroinflammatory miRNAs (mir-326, mir-155, and mir-27b) was increased and the relative expression of their target mRNA (PPARS, SOCS1, and CEBPA) was decreased in COVID-19 patients with increase of disease grade. A negative significant correlation was seen between each miRNA and its target mRNA. Based on bioinformatics analysis, some important pathways are affected by these pro-neuroinflammatory and anti-neuroinflammatory miRNAs, including PI3K-Akt, mRNA surveillance, mTOR, MAPK, Wnt, and AMPK signaling pathways. What we have found is that in patients with high severity of illness, the expression of pro-inflammatory miRNAs is increased, and conversely, the expression of anti-inflammatory miRNAs is decreased. Of course, it is clear that this situation follows a cytokine storm. Unfortunately, we have to say that this special condition not only causes serious damage to the brain but also causes damage to several organs and leads to multiple organ failure. We think that when immune cells are highly stimulated, cytokines and miRNAs can travel through the bloodstream to the whole body. This phenomenon has been mentioned by some researchers.15,16
Mir-155 is a central pro-inflammatory mediator in CNS by NF-κB dependent TLR signaling. It is synthesized inside macrophages and microglia.17–19mir-155 targets anti-inflammatory regulators such as SOCS1,17,19SHIP1,20C/EBP-β21 and IL13Rα1.22mir-155 inhibits the suppression of anti-inflammatory signaling and induces neuroinflammation. When mir-155 is expressed, it stimulates the transcription factor p53, and it targets the c-Maf transcription factor, which induces differentiation and inflammatory responses.23Mir-146a is an anti-inflammatory regulator in nerve cells, microglia, and astrocytes. It activates by NF-κB dependent TLR signaling.24,25 The Mir-146a targets MyD88 signaling complex, including IRAK1 and TRAF6, and acts as an NF-κB signaling regulator. In addition, Mir-146a targets other pro-inflammatory mediators including STAT-1,26,27IRF-527 and CFH.28,29 The polarization of macrophages and microglia are also altered by mir-146a.30mir-124 is also an anti-inflammatory miRNA and has a major role in neuronal differentiation31 and is highly expressed in microglia under normal conditions, but is not expressed in peripheral macrophages.32 Expression of mir-124 in microglia leads to anti-inflammatory effects33 by M2 phenotype.34 It is clear that mir-124 has anti-inflammatory activity by reducing inflammatory mediators and limiting microglia to activity. The role of mir-21 is very prominent in different types of CNS cells such as microglia35 and astrocytes,36 neurons,37 and oligodendrocytes.38Mir-21 is an anti-inflammatory regulator activated by TLR signaling. This induces the expression of the anti-inflammatory cytokine such as IL-10.39 In addition, mir-21 decreases TNF-α secretion in macrophages and microglia.40mir-27b targets an anti-inflammatory transcriptional activator, PPAR-γ; in human macrophages, this interaction blocks the induction of an anti-inflammatory phenotype. Inhibiting mir-27b also limits inflammatory signaling. It leads to produce inflammatory cytokines including IL-6 and TNF-α.41mir-326 is another pro-inflammatory miRNAs and can affect on differentiation of IL-17-producing Th17 cells. It was found that silencing mir-326 reduced EAE pathology.42 miRNAs have a cumulative effect on neuronal signaling and act together in inflammatory or anti-inflammatory pathways. For example, both mir-146a and mir-21 target different components of the TLR/MyD88/NF-κB and JAK-STAT pathways.26,28 In contrast, mir-155, mir-27b, and mir-326 activate the JAK-STAT pathway by targeting SOCS1 and SHIP1.19 It is interesting to note that miRNAs are also present in extracellular exosomes and can participate in intercellular communication.43 For example, mir-124, mir-21, and let-7 are found in exosomes and stimulate and regulate adjacent cells such as microglia and contribute to inflammatory signaling.44
One of main limitations of this study was to find and to collect COVID-19 patients with no comorbidities, no inflammatory autoimmune diseases, and no drug treatments. Theoretically, these factors can affect the expression of mRNAs and miRNAs. Second limitation was that we did not include COVID-19 patients caused by different variants of SARS-COV-2. Here, only COVID-19 patients with English variant (Lineage B.1.1.7) were included. We think that the expression of mRNAs and miRNAs may also be affected by virus variants. The third limitation was that we evaluated only 6 neuroinflammatory miRNAs in COVID-19 patients and it is suggested that other neuroinflammatory miRNAs could be studied in future studies.
ConclusionsThis study showed that the relative expression of anti-neuroinflammatory miRNAs (mir-21, mir-124, and mir-146a) was decreased and the relative expression of their mRNAs (IL-12p53, Stat3, and TRAF6) was increased in COVID-19 patients from asymptomatic to critical illness. Also, this study showed that the relative expression of pro-neuroinflammatory miRNAs (mir-326, mir-155, and mir-27b) was increased and the relative expression of their mRNA (PPARS, SOCS1, and CEBPA) was decreased in COVID-19 patients from asymptomatic to critical illness. A negative significant correlation was seen between mir-21 and IL-12p53 mRNA, mir-124 and Stat3, between mir-146a and TRAF6, between mir-27b and PPARS, between mir-155 and SOCS1, and between mir-326 and CEBPA mRNA in COVID-19 patients (P<0.05).
Authors’ contributions(I) Conception and design: R.K. and A.J., (II) Administrative support: R.K. and A.J., (III) Provision of study materials or patients: R.K., (IV) Collection and assembly of data: A.J., (V) Data analysis and interpretation: R.K. and A.J., (VI) Manuscript writing: All authors, (VII) Final approval of manuscript: All authors.
Ethics approval and consent to participate“The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.” All experiments were under the guidelines of the National Institute of Health, the provisions of the Declaration of Helsinki,and the ethics committee of Zahedan University of Medical Sciences, Zahedan, Iran. (Ethical code: IR.ZAUMS.REC.1399.317).
Consent for publicationNot.
Availability of data and materialNot.
FundingThis article was financially supported by Zahedan University of Medical Sciences, Zahedan, Iran (grant number: 9937).
Conflict of interestThere is no conflict of interest.
We thank the Reference Laboratory of Zahedan University of Medical Sciences.