This article provides an overview of stroke genetics studies ranging from the candidate gene approach to more recent studies by the genome wide association. It highlights the complexity of stroke owing to its different aetiopathogenic mechanisms, the difficulties in studying its genetic component, and the solutions provided to date. The study emphasises the importance of cooperation between the different centres, whether this takes place occasionally or through the creation of lasting consortiums. This strategy is currently essential to the completion of high-quality scientific studies that allow researchers to gain a better knowledge of the genetic component of stroke as it relates to aetiology, treatment, and prevention.
El presente artículo revisa la evolución de los estudios en genética del ictus desde la aproximación por gen candidato hasta los recientes estudios de genome wide association. Se destaca la complejidad de esta afección por sus muy variados mecanismos etiopatogénicos, las dificultades que comporta el estudio de su componente genético y las soluciones que se han aportado. Se subraya en especial el valor de las colaboraciones entre distintos centros, ya sea de manera puntual o sobre todo a través de la creación de consorcios estables. Esta estrategia actualmente se hace imprescindible a la hora de realizar estudios de alta calidad científica que permitan seguir avanzando en el conocimiento de las bases genéticas del ictus tanto en etiología, como en tratamiento y prevención.
In Spain, stroke is currently the second most frequent cause of death in the general population, as well as the leading cause of death in women (http://www.ine.es/prensa/np703.pdf). It is also the leading cause of disability in adults and the second most common cause of dementia.1
However, despite important advances in stroke prevention, diagnosis, and treatment in the past 2 decades, the World Health Organisation estimates that the incidence of this entity will increase by 27% by the year 2025.2 Researchers have described numerous risk factors associated with stroke, although in order to understand its causes it seems necessary to consider both external factors and the predisposition of each individual.3 The disease has been shown to have a hereditary component, and genetic load plays a very important role in its development.4 Technological advances in recent years have promoted genome-wide association studies (GWAS), which let researchers perform non-biased genetic analyses and discover new genes and metabolic pathways associated with stroke.
The purpose of this study is to review the current status and future perspectives of genetics in the field of cerebrovascular disease.
EvidenceGenetic factors in strokeUnlike monogenic disorders, complex diseases are characterised by their considerable aetiological and pathological diversity. Family history is recognised as one of the most important risk factors for many complex disorders; this is also the case for cardiovascular disease, cancer, or autoimmune disorders.5 In conclusion, even though these disorders are not monogenic or oligogenic, inherited genetic variations are apparent. Scientists believe that co-presence of different low-risk allelic variants whose combined risk is additive or multiplicative, and interaction of those variants with the environment, may play a crucial role in the development of complex disorders.5,6
Heritability rates between 30% and 50% have been described for ischaemic stroke.4 Because of its multiple causes, the numerous risk factors involved, and its array of different forms of presentation, stroke may be considered the final manifestation of any of a number of complex diseases. Apart from the simplest stroke classification scheme that distinguishes between ischaemic and haemorrhagic events, we can also identify several distinct aetiopathogenic subtypes. Several recently published studies have shown differences between the genetic heritability of lacunar, cardioembolic, and atherosclerotic strokes; the latter are the most strongly associated with family history.4,7,8
This also occurs in intracerebral haemorrhages (ICH), which account for 15% of all strokes. Pathogenic pathways involved in lobar haemorrhage, which is closely linked to cerebral amyloid angiopathy (CAA), may differ substantially from those involved in hypertensive haemorrhages.9,10
Therefore, genetic studies having to do with stroke should be designed so as to examine these particularities and accommodate the difficulties inherent to investigating heterogeneous diseases.
Genetic association studies in complex disorders: techniques and approachesResearch on most genetic causes of stroke, excluding those that follow classic Mendelian inheritance patterns, are generally based on case-control genetic association studies. These studies analyse the different frequencies of genetic variants in patients and healthy controls. Studying common variants or single nucleotide polymorphisms (SNPs) largely involves candidate-gene and genome-wide association, whereas the recently developed technique of complete exome sequencing lets scientists examine uncommon variants. Other techniques, such as analysis of copy-number variation or epigenome studies, will also be discussed below.
Candidate-gene studiesFor several decades, and until recently, the most common technique was the candidate-gene study. This approach consists of selecting genes that may be associated with a disease based on the pathophysiology of that disease and the pathways believed to be involved. Scientists identify previously described SNPs located near the candidate genes and select the most representative ones (tag SNPs) which will typically display linkage disequilibrium.11 These candidate tag SNPs are genotyped within a population and statistical analysis is used to determine if there are associations between genetic variants and the clinical spectrum.
These studies present several problems. On the one hand, the fact that these genes are selected based on a priori hypotheses may mean that some researchers look for spurious differences based on preconceived notions. Meanwhile, they may fail to analyse genes that are potentially involved in the disease.
Multiple genes have been linked to increased risk of ischaemic stroke by means of candidate gene studies. However, a recent study showed that none of these potential associations were statistically significant once a statistical correction had been applied to adjust for the number of SNPs analysed.4 Furthermore, no results obtained using this technique have ever been clearly replicated, which shows that the approach is inconsistent and unreliable where ischaemic stroke is concerned.4,12
Even so, this technique has shown that mutations in the COL4A1 and COL4A2 genes are associated with ICH. Heterotrimers formed by collagen IV alpha 1 (COL4A1) and alpha 2 (COL4A2) are one of the main components of nearly all basement membranes of the body, and they have been associated with numerous diseases that may be ocular, renal, or muscular as well as cerebrovascular.13 Initially, doctors using familial inheritance studies found that mutations in COL4A1 caused porencephaly type I, and this condition was later shown to cause both sporadic and recurrent ICH, as well as lacunar stroke and leukoaraiosis.14–16
A recent study, also related to ICH, showed that patients bearing haplotypes ¿2 and ¿4 of the APOE gene are at higher risk for lobar ICH (odds ratio [OR] 1.82 [95% CI, 1.50-2.23]; P=6.6×10−10 for ¿2 and OR 2.20 [95% CI, 1.85-2.63]; P=2.4×10−11, for ¿4). This is probably due to the effect of these variants on the risk of developing CAA.17 The same group later demonstrated that the APOE*E2 variant was associated with a greater volume of ICH, and consequently with poorer functional prognosis and higher mortality rate.18
GWASGWAS, which let researchers measure the allele frequency of hundreds of thousands of SNPs in cases versus controls, were developed for 2 main reasons. First came the creation of the International HapMap Project which determined the characteristics, linkage equilibrium, and allele frequencies of SNPs found in the genome of 270 individuals representing 4 different ancestral origins. Its purpose was to create a public genome-wide database that could be freely accessed by all researchers.5,19 Secondly, significant technological advances and platforms were developed that permitted genotyping 500000 to 2.7 million SNPs per individual at consistently lower costs. GWAS studies constitute a much more exhaustive means of studying the genome than candidate studies.20 They are not affected by preconceived hypothesis biases and may also let scientists identify new associations even when the gene has not yet been linked to the disease, unlike earlier approaches. Nevertheless, GWAS has other types of limitations. Its main 2 limitations are the large sample sizes that are needed and the difficulty of interpreting potential positive results.21 These studies also require strict quality control processes to detect genotyping errors and assess possible stratification in populations.22 The latter limitation arises because the allele frequencies of an SNP vary according to each individual's ethnic makeup, even between individuals sharing an ethnic phenotype. Since the above situation may give rise to false results if the case and control populations do not have comparable genetic origins, the pertinent adjustments have to be made. In fact, the best way of eliminating these inconsistencies is by replicating results obtained from very large independent samples.23 Considering the large numbers of analyses that must be performed (at least one per genotyped SNP), statistical penalties should be applied for multiple comparisons such that the significance level (P-value) may be set at 10−8 (in cases of analysing one million SNPs).6 If studies are to reach the necessary statistical power, their sample sizes must be increased by a significant amount.
The first results having to do with complex diseases were published in 2005, when complement factor H was found to be involved in age-related macular degeneration.24 Interesting discoveries have since been made in the areas of type 1 diabetes mellitus (PPARG25, KCNJ11,26 and TCF7L227); rheumatoid arthritis and type 2 diabetes mellitus (PTPN2228,29) and Crohn disease (NOD2)30 and many others. Large numbers of loci have been found to be implicated in the predisposition or risk of developing the above diseases.
In contrast with the discouraging results that have been delivered by other approaches, several associations with ischaemic stroke have been replicated consistently. Such is the case of associations discovered through GWAS for other diseases associated with ischaemic stroke and which have also been determined to be stroke risk factors.31 As such, 2 variants, rs1906591 (OR 1.48 [95% CI, 1.28-1.71]; P=4.7×10−6) and rs7193343 (OR 1.22 [95% CI, 1.10-1.35]; P=.00021), in chromosomes 4q25 and 16q22 in regions near genes PITX2 and ZFHX3 respectively, were initially identified as contributing to the risk of developing atrial fibrillation. Since then, they have also been associated with higher risk of presenting a cardioembolic stroke.32,33
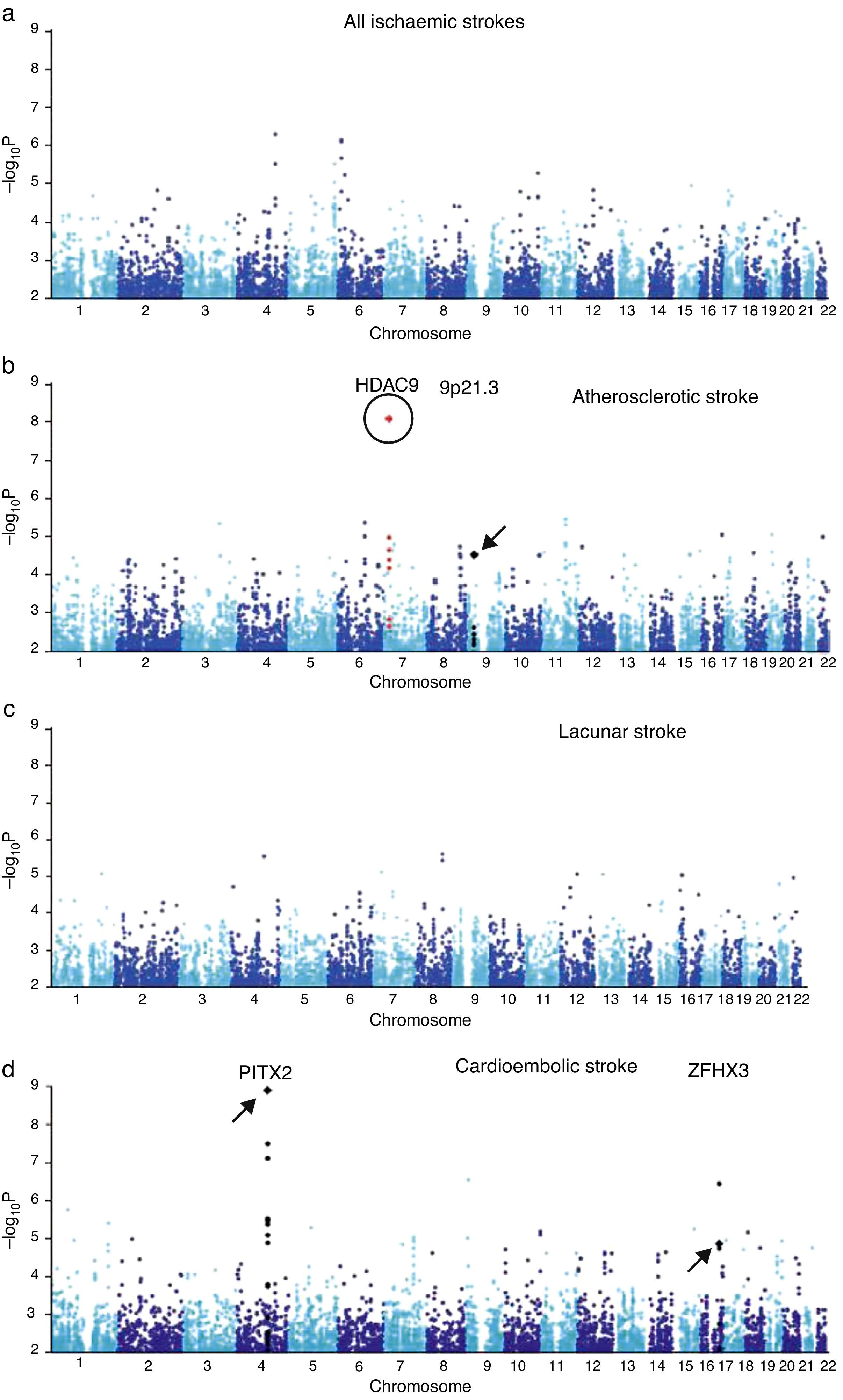
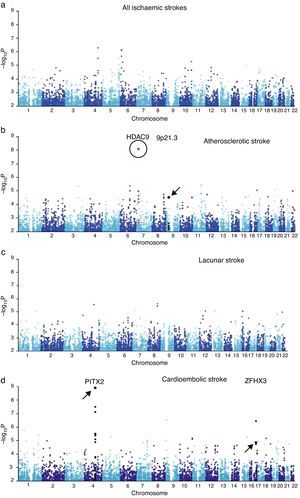
A region in chromosome 9p21 near the CDKN2A/B genes has been linked to myocardial infarction and coronary artery disease. A later candidate gene study of these loci using a candidate gene found an association between the SNP rs1537378 (OR 1.21; [95% CI, 1.07-1.37]; P=.002) and atherothrombotic stroke.34 GWAS has also identified a new association with atherothrombotic stroke: polymorphism rs11984041 (OR 1.42 [95% CI, 1.28-1.57]; P=1.87×10−11). This variant is located in the region of gene HDAC9 on chromosome 7p21.1, but its pathogenic mechanism is unknown (Fig. 1).35
GWAS results from samples from the UK and Germany. (a) Results from all ischaemic strokes. (b) Atherosclerotic stroke. (c) Lacunar stroke. (d) Cardioembolic stroke. The circle indicates the new locus in HDAC9. Loci described in earlier literature are indicated with arrows.
Published with permission from Macmillan Publishers Ltd: Nature Genetics, 2012.35
Adapted with permission from Macmillan Publishers Ltd: Nature Genetics, 2012.35
Regarding new associations, a Japanese team has identified a significant link between a polymorphism of the PRKCH gene, rs2230500 on chromosome 14 (OR 1.66 [95% CI, 1.33-2.09]; P=9.84×10−6), with lacunar stroke. Replication results have been dissimilar in other ethnic groups.4,36
Exome sequencingOne of the weak points of GWAS is that it only analyses common variants (population frequency>1%) without considering rare variants. There may in fact be several rare variants that contribute to genetic risk of complex diseases. In order to identify them, scientists are developing new exome sequencing techniques. These new techniques identify all polymorphisms or mutations found in protein-coding regions of the genome. As such, they are able to detect the rare variants that cannot be identified by GWAS. While this approach is promising, its methodology is still being developed and doctors have yet to agree on the best way to manage the huge amounts of data generated by these analyses.37
Analysis of copy-number variationCopy-number variation (CNV) is a structural variation of the genome that may occur as (micro) deletions, duplications, or inversions, creating an anomalous number of copies of a DNA fragment. This variation may affect both the expression and the makeup of proteins that are coded in this region.38–40 At present, very little has been published regarding stroke. While one study of patients with ischaemic stroke did not detect associated CNV, the sample size was small and the different stroke subtypes were not taken into account.41 However, there are genome-wide studies currently underway that include analysis of multiple CNVs in their analysis platforms.
EpigeneticsAnother emerging field is epigenetics, a discipline that studies heritable changes in gene regulation that do not affect the DNA sequence. Although all the cells in our body display identical genomes, genetic transcription in each tissue type is regulated by different epigenetic mechanisms. Chief among these mechanisms are DNA methylation, and modifications to histones and non-coding RNA.42 Of these mechanisms, DNA methylation is the most studied and best understood to date. The most important findings were obtained from epigenetic studies in tumour tissue. For example, methylation of the MGMT gene inactivates enzyme O (6)-methylguanine-DNA methyltransferase in cases of glioblastoma multiforme, which is associated with a better response to chemotherapy with temozolamide. This finding is already being applied in clinical practice.43 Epigenetic studies are currently being expanded to contemplate other diseases, including stroke. One describes an association between stroke and overall DNA hypomethylation, and that study is currently being replicated.44 Within the framework of the Spanish Consortium on Stroke Genetics, several studies in this field are currently underway and results are not yet available.
Risk scales and genetic networksDesigning genetic scores and genetic networks is becoming increasingly relevant for purposes of integrating the information and predictive ability of each SNP associated with a phenotype. The most commonly used methods for creating genetic scores are summing risk alleles (1 point per allele) or calculating weighted risk (the score for each allele depends on the OR reported for that allele). A very recent study showed an association between the score calculated based on risk alleles for arterial hypertension and the risk of experiencing deep ICH.45
One way of organising knowledge is by creating networks or algorithms.46 One example is the Bayesian network, a tool in statistics that represents an array of associated variables set out according to the conditional dependencies that have been established between them. These models let researchers estimate the probability of the appearance of latent variables based on known probabilities. The main advantages of Bayesian networks are that they permit bidirectional interference (that is, effect-to-cause and cause-to-effect) as well as rapid updating of knowledge.47
Genomic convergenceThe concept of genomic convergence is becoming increasingly relevant to genetic studies of complex diseases. It is based on combining and unifying the information and data provided by each different type of study to achieve a greater capacity for discovery and more robust results.48 Several polymorphisms of gene TTC7B were recently shown to confer susceptibility to ischaemic stroke.49
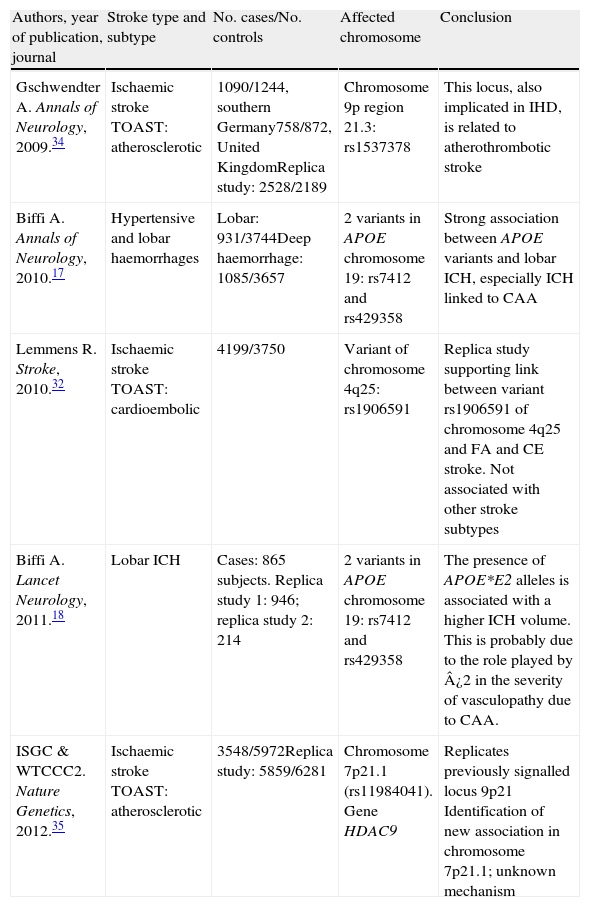
The creation of consortia: GeneStroke, the Spanish consortium on stroke geneticsFollowing the creation of large international consortia, such as the UK's Wellcome Trust Case Control Consortium, the International Stroke Genetics Consortium (ISGC) was formed in 2007 (http://www.strokegenetics.org). The ISGC's activities have included identifying and fostering numerous associations (Table 1) and many more ambitious international projects in this field are currently underway.
Main articles completed within the framework of the International Stroke Genetics Consortium and published in major journals
| Authors, year of publication, journal | Stroke type and subtype | No. cases/No. controls | Affected chromosome | Conclusion |
| Gschwendter A. Annals of Neurology, 2009.34 | Ischaemic stroke TOAST: atherosclerotic | 1090/1244, southern Germany758/872, United KingdomReplica study: 2528/2189 | Chromosome 9p region 21.3: rs1537378 | This locus, also implicated in IHD, is related to atherothrombotic stroke |
| Biffi A. Annals of Neurology, 2010.17 | Hypertensive and lobar haemorrhages | Lobar: 931/3744Deep haemorrhage: 1085/3657 | 2 variants in APOE chromosome 19: rs7412 and rs429358 | Strong association between APOE variants and lobar ICH, especially ICH linked to CAA |
| Lemmens R. Stroke, 2010.32 | Ischaemic stroke TOAST: cardioembolic | 4199/3750 | Variant of chromosome 4q25: rs1906591 | Replica study supporting link between variant rs1906591 of chromosome 4q25 and FA and CE stroke. Not associated with other stroke subtypes |
| Biffi A. Lancet Neurology, 2011.18 | Lobar ICH | Cases: 865 subjects. Replica study 1: 946; replica study 2: 214 | 2 variants in APOE chromosome 19: rs7412 and rs429358 | The presence of APOE*E2 alleles is associated with a higher ICH volume. This is probably due to the role played by ¿2 in the severity of vasculopathy due to CAA. |
| ISGC & WTCCC2. Nature Genetics, 2012.35 | Ischaemic stroke TOAST: atherosclerotic | 3548/5972Replica study: 5859/6281 | Chromosome 7p21.1 (rs11984041). Gene HDAC9 | Replicates previously signalled locus 9p21 Identification of new association in chromosome 7p21.1; unknown mechanism |
CAA: cerebral amyloid angiopathy; CE: cardioembolic; IHD: ischaemic heart disease; AF: atrial fibrillation; ICH: intracerebral haemorrhage; TOAST: Trial of Org 10172 in Acute Stroke Treatment.
GeneStroke, the Spanish Stroke Genetics Consortium (http://www.genestroke.com), was formed in 2009 for the main purpose of studying the genetic basis of stroke and promoting interaction between groups with common interests in this field. GeneStroke has launched a number of multi-centre cooperative projects related to stroke aetiology, progression, and prognosis. Further information about these projects and participants is available on the website (www.genestroke.com).
The Spanish Stroke Genetics Consortium promotes a model of large collaborative studies that are able to meet the challenges faced by research in the area of cerebrovascular disease genetics.
ConclusionsStroke is one of the most prevalent diseases in our setting and it has a major impact in terms of morbidity and mortality. Its heritability is high, and a better knowledge of its genetic basis may contribute to identifying at-risk individuals and designing more efficient treatments and prevention strategies. The study of the genetic substrate should be a priority area, along with cooperation between different groups and professionals striving to complete high-quality scientific projects that may benefit society.
Conflicts of interestThe authors have no conflicts of interest to declare.
We would first like to thank all permanent members of the Spanish Stroke Genetics Consortium (GeneStroke) and any other members who have made sporadic contributions to these projects. We would also like to express our particular gratitude to the members of the International Stroke Genetics Consortium.
Please cite this article as: Giralt-Steinhauer E, Jiménez-Conde J, Soriano Tárraga C, Mola M, Rodríguez-Campello A, Cuadrado-Godia E, et al. Aproximación al conocimiento de las bases genéticas del ictus. Consorcio español de genética del ictus. Neurología. 2014;29:560–566.