Epigenetics alterations, including aberrant DNA methylation, have been associated with gastric carcinogenesis. Single nucleotide polymorphisms (SNPs) in DNA methyltransferases (DNMTs) may influence protein expression and therefore affect DNA regulation and susceptibility for Gastric Cancer (GC).
We have performed a systematic review and meta-analysis involving 11 studies and a total of 24 SNPs in DNMTs were analyzed. According to literature, only 4 SNPs, DNMT1 rs16999593, DNMT2 rs11254413 and DNMT3A rs7560488 and DNMT3A rs36012910, were associated with GC. DNMT1 rs16999593 and DNMT3A rs7560488C allele and DNMT3A rs36012910 G allele showed an increased risk for GC. On the other hand, DNMT2 rs11254413 G allele presented a protective effect for GC. Additionally, the meta-analysis evaluated the SNPs analyzed in more than one study (n=6). Results revealed that only DNMT1 rs16999593 had a statistically significant association with GC development (OR=1.31; 95% CI=1.08–1.60; p=0.006 for TC+CC genotypes).
Our study suggests that DNMT2 rs11254413, DNMT3A rs7560488, DNMT3A rs36012910 and, specially, DNMT1 rs16999593 may have an association with GC development. Nevertheless, further studies are need using different populations to clarify this association with GC risk.
Epigenetics, and more extensively DNA methylation, has been studied in the past decades and associated with several diseases, including cancer. Literature shows that altered epigenetic control of gene expression has an important role in carcinogenesis.1 DNA methylation is an important epigenetic mark in tumorigenesis, in which a methyl group is added to the 5′ position of the cytosine residue in a CpG dinucleotide (CpG islands, CGI), in the 5′-flanking promoter's genes.2 Aberrant methylation of promoter's genes is an important hallmark of cancer cells and, in Gastric Cancer (GC), a large number of genes involved in carcinogenesis and clinical outcome accumulate aberrant methylation. Promoter methylation is an important mechanism of inactivation of tumour suppressor genes, in cancer cells.2
The process of promoter methylation is catalyzed by DNA methyltransferases (DNMTs) which organize, regulate and maintain properly mammalian genomes.3 Literature describes three types of DNMTs: DNMT1, over-expressed in human cancers including GC, that catalyze post-replication DNA methylation and maintain the methylation patterns during cell divisions4–6; DNMT2, which is the most conserved and has a role in both DNA and RNA methylation7; and DNMT3, which is divided into two types (DNMT3a and DNMT3b) that are responsible for de novo methylation during gametogenesis and embryogenesis, and that also seem to be over-expressed in human cancers, including GC.3,6 Recently, literature has suggested that single nucleotide polymorphisms (SNPs) in DNMTs could promote functional consequences affecting methylation and therefore individual's susceptibility to cancer development.6,8
Several studies have reported a potential association between SNPs in DNMTs and the GC susceptibility.3,9–11 Nevertheless, the observed associations of these studies are inconsistent and some studies are not large enough to take conclusions on the effect of DNMTs SNPs on GC. As a result, we have performed a systematic review and meta-analysis of all eligible studies to understand the possible association of SNPs in DNMTs genes and GC susceptibility.
Materials and methodsSearch strategyThe present systematic review and meta-analysis followed PRISMA guidelines for Systematic Review and Meta-analyses. Literature was searched on PubMed data base until March 2016 applying the following query: “dna modification methylases”[MeSH Terms] OR (“dna”[All Fields] AND “modification”[All Fields] AND “methylases”[All Fields]) OR “dna modification methylases”[All Fields] OR (“dna”[All Fields] AND “methyltransferase”[All Fields]) OR “dna methyltransferase”[All Fields] OR DNMT[All Fields]) AND (“polymorphism, genetic”[MeSH Terms] OR (“polymorphism”[All Fields] AND “genetic”[All Fields]) OR “genetic polymorphism”[All Fields] OR “polymorphism”[All Fields]) AND (“stomach neoplasms”[MeSH Terms] OR (“stomach”[All Fields] AND “neoplasms”[All Fields]) OR “stomach neoplasms”[All Fields] OR (“gastric”[All Fields] AND “cancer”[All Fields]) OR “gastric cancer”[All Fields].
Inclusion and exclusion criteriaThe literature search was limited to original studies performed in humans and no publication year restriction was applied. Only case–control studies with histologically-confirmed adenocarcinoma cases were included in this analysis. All DNMTs polymorphisms were selected for the analysis. Regarding the language of the papers, only studies written in English, Portuguese or Spanish were selected. Reviews, Meta-analysis, Systematic Reviews and studies not related to gastric cancer were excluded from this analysis.
Data extractionTwo authors performed the data extraction and all disagreements were resolved with the opinion of a third author. For each study, the following items were collected: country, type of study, ethnicity, matching criteria, age (years), polymorphisms, genotype method, and the number of cases and controls. Data concerning genotype distribution and the relative risk were also extracted for all polymorphisms included in the studies.
Statistical analysisThe evidence of Hardy Weinberg Equilibrium (HWE) in controls was recalculated in the present meta-analysis through the application of the online software (http://www.had2know.com/academics/hardy-weinberg-equilibrium-calculator-2-alleles.html). A p-value less than 0.05 of HWE was considered significant.
Meta-analysis for DNMTs SNPs was conducted by using the software Review Manager, version 5.3.5 (Copenhagen: The Nordic Cochrane Centre, The Cochrane Collaboration, 2014). The calculated OR (95% CI) was performed with Mantel–Haenszel statistical method and Random effects analysis model. A p-value less than 0.05 was considered significant.
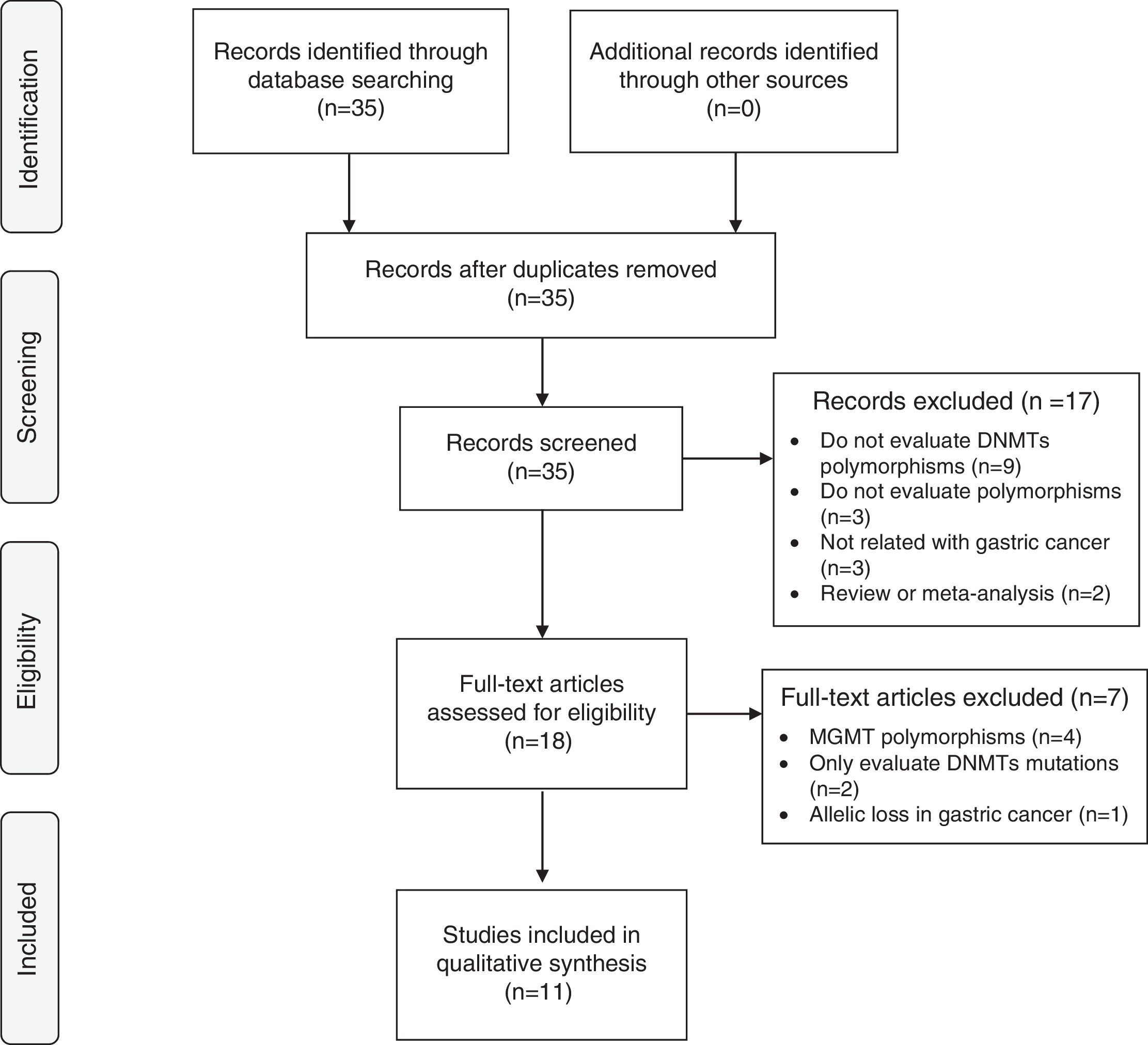
ResultsLiterature searchA total of 35 papers were evaluated from which only 11 matched inclusion criteria. Seventeen articles were excluded due to the exclusion criteria by screening the titles and abstracts. After a comprehensive evaluation of the remaining 18 articles, a total of 7 studies were excluded: 4 articles did not evaluate polymorphisms of DNMTs, only O6-methylguanine-DNA methyltransferase (MGMT)12–15; 2 articles only reported mutations on DNMTs5,16; 1 study was about allelic loss in gastric cancer.17 Finally, 11 studies involving a total of 3049 cases and 5185 controls were included in this analysis – Fig. 1.
Flow chart of studies identification, exclusion and inclusion. Adapted from: Moher D, Liberati A, Tetzlaff J, Altman DG, The PRISMA Group (2009). Preferred Reporting Items for Systematic Reviews and Meta-analyses: The PRISMA Statement. PLoS Med 6(6): e1000097. doi:10.1371/journal.pmed1000097.
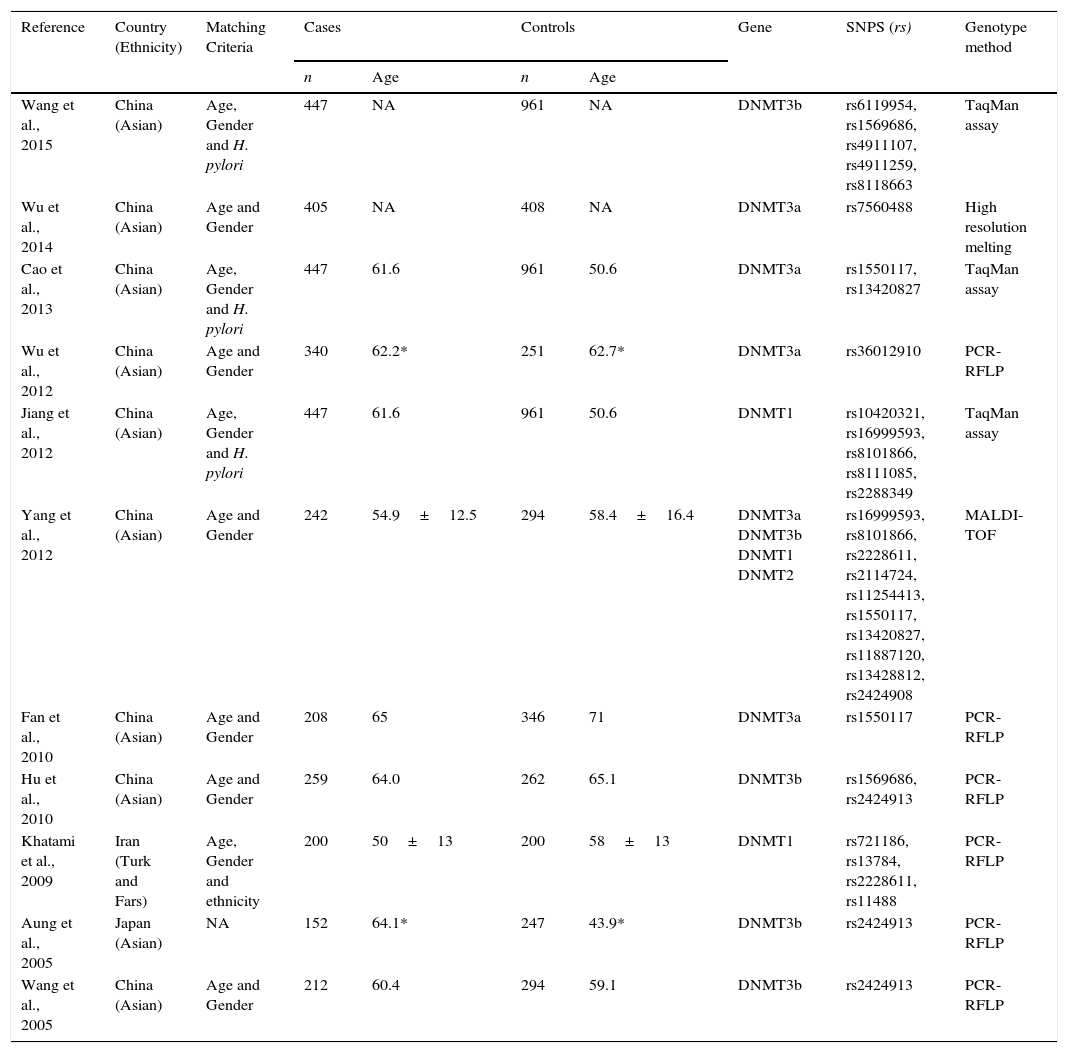
Table 1 describes the principal baseline characteristics of included studies. The majority of the studies were performed in China,3,8–11,18–21 one study in Iran22 and one in Japan.23 Almost all studies have selected the controls and cases using age and gender as matching criteria, but some studies also used Helicobacter pylori infection status and Ethnicity. The number of cases and controls varies between the studies. A total of 24 polymorphisms were evaluated, 10 of DNMT1, 1 of DNMT2, 6 of DNMT3A and 7 of DNMT3B. The majority of studies were performed using Polymerase Chain Reaction–Restriction Fragment Length Polymorphism (PCR-RFLP) methodology, but other methods of genotyping were used like TaqMan assay, High Resolution Melting (HRM) and Matrix Assisted Laser Desorption/Ionization – Time of Flight (MALDI-TOF). Supplementary Table 1 shows the characteristics of all polymorphisms included in the studies. The re-calculated HWE for controls is also shown in supplementary Table 2.
Characteristics of all studies included in the analysis.
| Reference | Country (Ethnicity) | Matching Criteria | Cases | Controls | Gene | SNPS (rs) | Genotype method | ||
|---|---|---|---|---|---|---|---|---|---|
| n | Age | n | Age | ||||||
| Wang et al., 2015 | China (Asian) | Age, Gender and H. pylori | 447 | NA | 961 | NA | DNMT3b | rs6119954, rs1569686, rs4911107, rs4911259, rs8118663 | TaqMan assay |
| Wu et al., 2014 | China (Asian) | Age and Gender | 405 | NA | 408 | NA | DNMT3a | rs7560488 | High resolution melting |
| Cao et al., 2013 | China (Asian) | Age, Gender and H. pylori | 447 | 61.6 | 961 | 50.6 | DNMT3a | rs1550117, rs13420827 | TaqMan assay |
| Wu et al., 2012 | China (Asian) | Age and Gender | 340 | 62.2* | 251 | 62.7* | DNMT3a | rs36012910 | PCR-RFLP |
| Jiang et al., 2012 | China (Asian) | Age, Gender and H. pylori | 447 | 61.6 | 961 | 50.6 | DNMT1 | rs10420321, rs16999593, rs8101866, rs8111085, rs2288349 | TaqMan assay |
| Yang et al., 2012 | China (Asian) | Age and Gender | 242 | 54.9±12.5 | 294 | 58.4±16.4 | DNMT3a DNMT3b DNMT1 DNMT2 | rs16999593, rs8101866, rs2228611, rs2114724, rs11254413, rs1550117, rs13420827, rs11887120, rs13428812, rs2424908 | MALDI-TOF |
| Fan et al., 2010 | China (Asian) | Age and Gender | 208 | 65 | 346 | 71 | DNMT3a | rs1550117 | PCR-RFLP |
| Hu et al., 2010 | China (Asian) | Age and Gender | 259 | 64.0 | 262 | 65.1 | DNMT3b | rs1569686, rs2424913 | PCR-RFLP |
| Khatami et al., 2009 | Iran (Turk and Fars) | Age, Gender and ethnicity | 200 | 50±13 | 200 | 58±13 | DNMT1 | rs721186, rs13784, rs2228611, rs11488 | PCR-RFLP |
| Aung et al., 2005 | Japan (Asian) | NA | 152 | 64.1* | 247 | 43.9* | DNMT3b | rs2424913 | PCR-RFLP |
| Wang et al., 2005 | China (Asian) | Age and Gender | 212 | 60.4 | 294 | 59.1 | DNMT3b | rs2424913 | PCR-RFLP |
NA, not available; * Median (range); H. pylori – Helicobacter pylori; DNMT1 – DNA (cytosine-5)-methyltransferase 1; DNMT2 – TRNA (cytosine38-C5)-methyltransferase; DNMT3a – DNA (cytosine-5)-methyltransferase 3 alpha; DNMT3b – DNA (cytosine-5-)-methyltransferase 3 beta; MALDI-TOF – Matrix-Assisted Laser Desorption/Ionization-Time of Flight; PCR-RFLP – Polymerase Chain Reaction-Restriction Fragment Length Polymorphism.
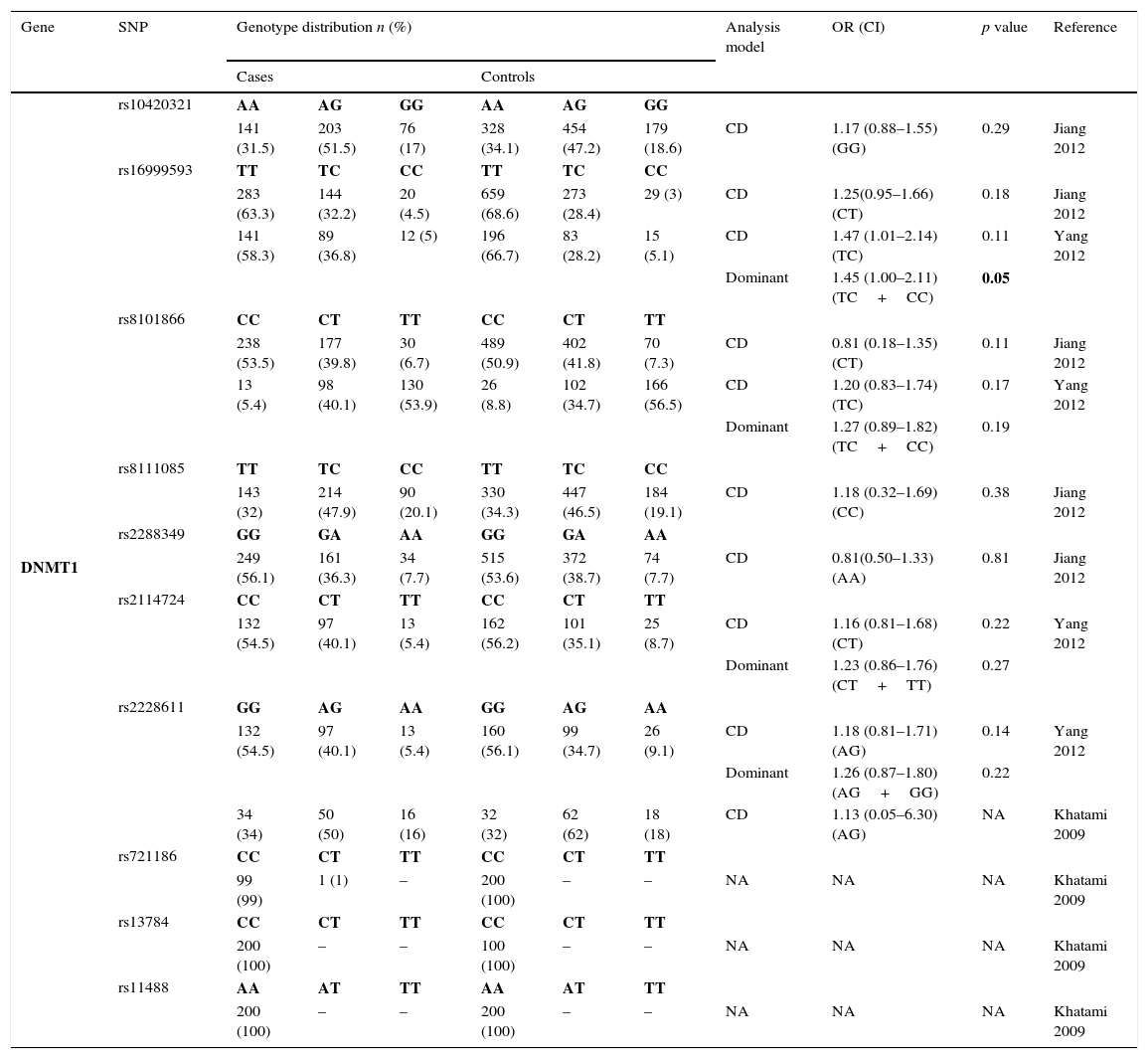
The genotyping results of all studies are described in Table 2. A total of 10 SNPs in DNMT1 were studied by 3 different studies and only rs16999593, rs8101866 and rs2228611 have data from more than one population3,11,22 – Table 2.1. The risk analysis revealed that only one study reported a significant risk association (rs16999593) with GC development (OR=1.45; 95% CI=1.00–2.11; p=0.05, for C allele).
Genotyping results for DNMT1 of all studies included.
| Gene | SNP | Genotype distribution n (%) | Analysis model | OR (CI) | p value | Reference | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Cases | Controls | ||||||||||
| DNMT1 | rs10420321 | AA | AG | GG | AA | AG | GG | ||||
| 141 (31.5) | 203 (51.5) | 76 (17) | 328 (34.1) | 454 (47.2) | 179 (18.6) | CD | 1.17 (0.88–1.55) (GG) | 0.29 | Jiang 2012 | ||
| rs16999593 | TT | TC | CC | TT | TC | CC | |||||
| 283 (63.3) | 144 (32.2) | 20 (4.5) | 659 (68.6) | 273 (28.4) | 29 (3) | CD | 1.25(0.95–1.66) (CT) | 0.18 | Jiang 2012 | ||
| 141 (58.3) | 89 (36.8) | 12 (5) | 196 (66.7) | 83 (28.2) | 15 (5.1) | CD | 1.47 (1.01–2.14) (TC) | 0.11 | Yang 2012 | ||
| Dominant | 1.45 (1.00–2.11) (TC+CC) | 0.05 | |||||||||
| rs8101866 | CC | CT | TT | CC | CT | TT | |||||
| 238 (53.5) | 177 (39.8) | 30 (6.7) | 489 (50.9) | 402 (41.8) | 70 (7.3) | CD | 0.81 (0.18–1.35) (CT) | 0.11 | Jiang 2012 | ||
| 13 (5.4) | 98 (40.1) | 130 (53.9) | 26 (8.8) | 102 (34.7) | 166 (56.5) | CD | 1.20 (0.83–1.74) (TC) | 0.17 | Yang 2012 | ||
| Dominant | 1.27 (0.89–1.82) (TC+CC) | 0.19 | |||||||||
| rs8111085 | TT | TC | CC | TT | TC | CC | |||||
| 143 (32) | 214 (47.9) | 90 (20.1) | 330 (34.3) | 447 (46.5) | 184 (19.1) | CD | 1.18 (0.32–1.69) (CC) | 0.38 | Jiang 2012 | ||
| rs2288349 | GG | GA | AA | GG | GA | AA | |||||
| 249 (56.1) | 161 (36.3) | 34 (7.7) | 515 (53.6) | 372 (38.7) | 74 (7.7) | CD | 0.81(0.50–1.33) (AA) | 0.81 | Jiang 2012 | ||
| rs2114724 | CC | CT | TT | CC | CT | TT | |||||
| 132 (54.5) | 97 (40.1) | 13 (5.4) | 162 (56.2) | 101 (35.1) | 25 (8.7) | CD | 1.16 (0.81–1.68) (CT) | 0.22 | Yang 2012 | ||
| Dominant | 1.23 (0.86–1.76) (CT+TT) | 0.27 | |||||||||
| rs2228611 | GG | AG | AA | GG | AG | AA | |||||
| 132 (54.5) | 97 (40.1) | 13 (5.4) | 160 (56.1) | 99 (34.7) | 26 (9.1) | CD | 1.18 (0.81–1.71) (AG) | 0.14 | Yang 2012 | ||
| Dominant | 1.26 (0.87–1.80) (AG+GG) | 0.22 | |||||||||
| 34 (34) | 50 (50) | 16 (16) | 32 (32) | 62 (62) | 18 (18) | CD | 1.13 (0.05–6.30) (AG) | NA | Khatami 2009 | ||
| rs721186 | CC | CT | TT | CC | CT | TT | |||||
| 99 (99) | 1 (1) | – | 200 (100) | – | – | NA | NA | NA | Khatami 2009 | ||
| rs13784 | CC | CT | TT | CC | CT | TT | |||||
| 200 (100) | – | – | 100 (100) | – | – | NA | NA | NA | Khatami 2009 | ||
| rs11488 | AA | AT | TT | AA | AT | TT | |||||
| 200 (100) | – | – | 200 (100) | – | – | NA | NA | NA | Khatami 2009 | ||
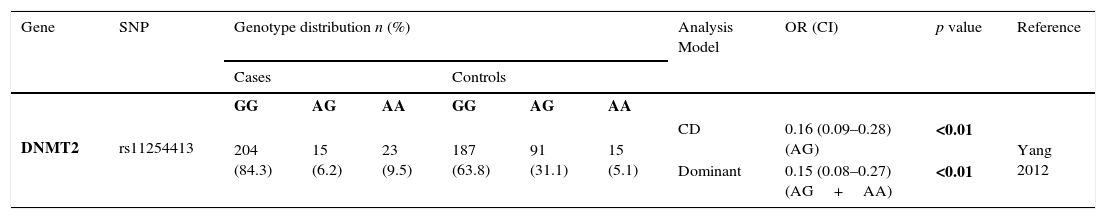
Only one study evaluated SNPs in DNMT2 and GC, revealing that rs112254413 A allele was associated with a protection for the development of GC (OR=0.15; 95% CI=0.08–0.27; p<0.01)3 – Table 2.2.
Genotyping results for DNMT2 of all studies included.
| Gene | SNP | Genotype distribution n (%) | Analysis Model | OR (CI) | p value | Reference | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Cases | Controls | ||||||||||
| DNMT2 | rs11254413 | GG | AG | AA | GG | AG | AA | ||||
| 204 (84.3) | 15 (6.2) | 23 (9.5) | 187 (63.8) | 91 (31.1) | 15 (5.1) | CD | 0.16 (0.09–0.28) (AG) | <0.01 | Yang 2012 | ||
| Dominant | 0.15 (0.08–0.27) (AG+AA) | <0.01 | |||||||||
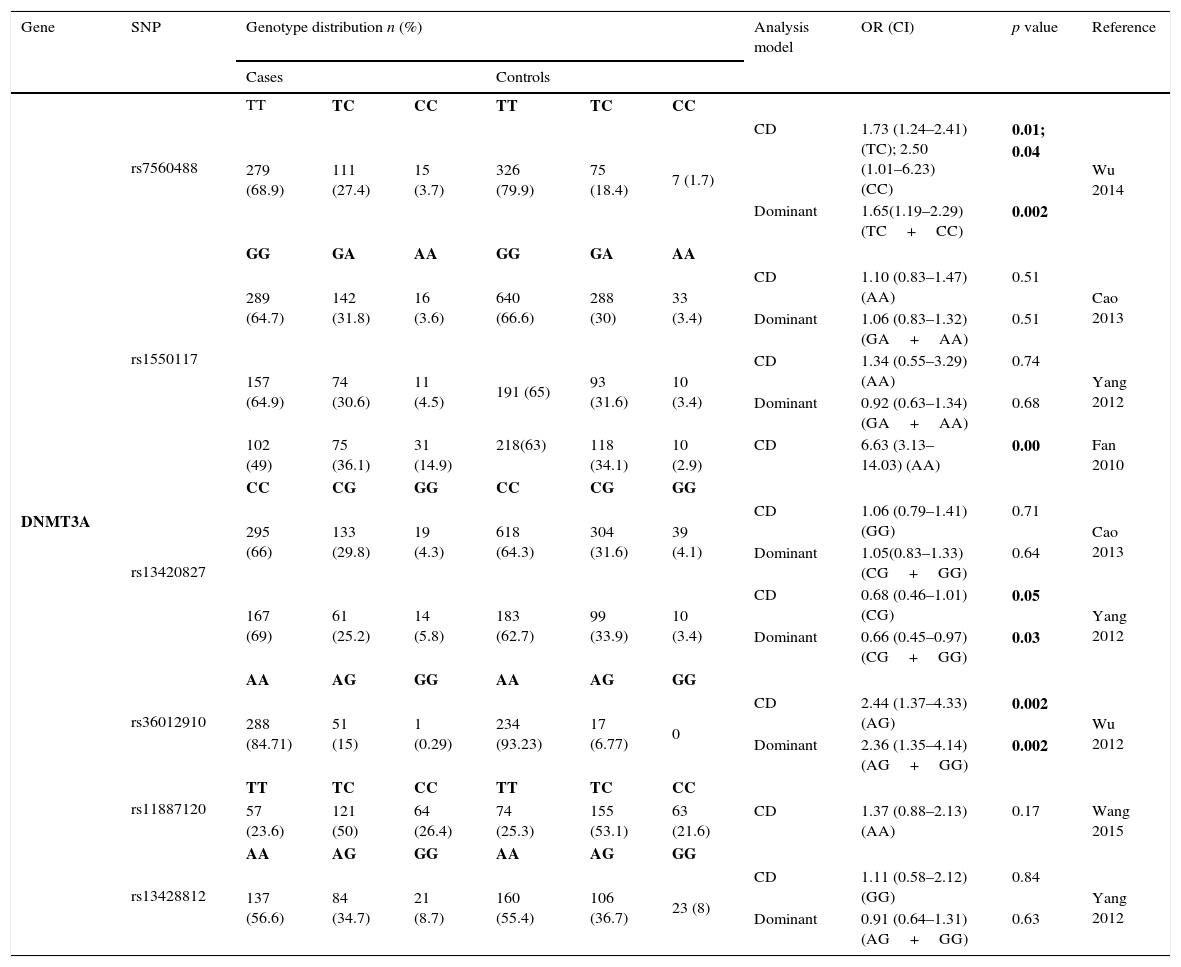
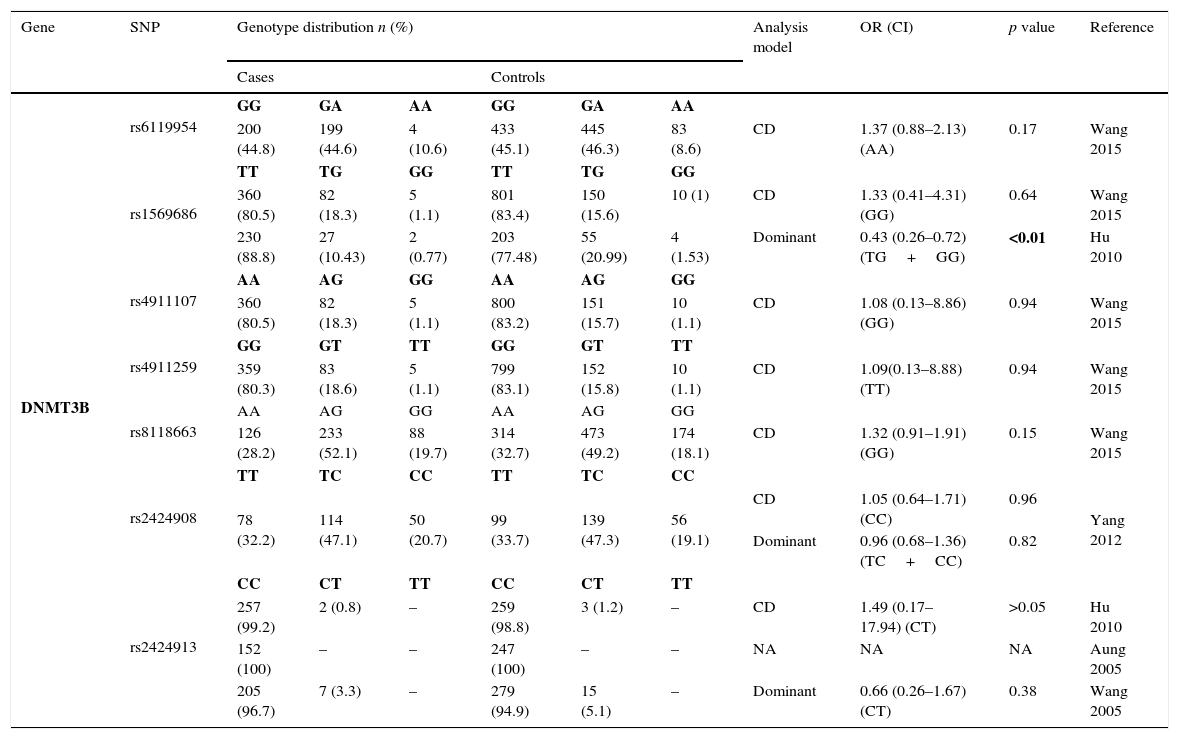
Thirteen SNPs of DNMT3 (6 from DNMT3A and 7 from DNMT3B) were evaluated by 9 different studies3,8–10,19–21,23,24 – Tables 2.3 and 2.4. The risk analysis have shown that DNMT3A rs7560488 (OR=1.65; 95%CI=1.19–2.29; p=0.002, for C allele) and rs36012910 (OR=2.36; 95%CI=1.35–4.14; p=0.002, for allele G) seem to be associated with an increased risk for GC.8,19 On the other hand, DNMT3A rs1550117 and rs13420867 show contradictory findings between the reported studies. In DNMT3B, only rs1569686 showed a statistically significant association (p<0.01) with GC, nevertheless the results are contradictory among the different studies.
Genotyping results for DNMT3A of all studies included.
| Gene | SNP | Genotype distribution n (%) | Analysis model | OR (CI) | p value | Reference | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Cases | Controls | ||||||||||
| DNMT3A | rs7560488 | TT | TC | CC | TT | TC | CC | ||||
| 279 (68.9) | 111 (27.4) | 15 (3.7) | 326 (79.9) | 75 (18.4) | 7 (1.7) | CD | 1.73 (1.24–2.41) (TC); 2.50 (1.01–6.23) (CC) | 0.01; 0.04 | Wu 2014 | ||
| Dominant | 1.65(1.19–2.29) (TC+CC) | 0.002 | |||||||||
| rs1550117 | GG | GA | AA | GG | GA | AA | |||||
| 289 (64.7) | 142 (31.8) | 16 (3.6) | 640 (66.6) | 288 (30) | 33 (3.4) | CD | 1.10 (0.83–1.47) (AA) | 0.51 | Cao 2013 | ||
| Dominant | 1.06 (0.83–1.32) (GA+AA) | 0.51 | |||||||||
| 157 (64.9) | 74 (30.6) | 11 (4.5) | 191 (65) | 93 (31.6) | 10 (3.4) | CD | 1.34 (0.55–3.29) (AA) | 0.74 | Yang 2012 | ||
| Dominant | 0.92 (0.63–1.34) (GA+AA) | 0.68 | |||||||||
| 102 (49) | 75 (36.1) | 31 (14.9) | 218(63) | 118 (34.1) | 10 (2.9) | CD | 6.63 (3.13–14.03) (AA) | 0.00 | Fan 2010 | ||
| rs13420827 | CC | CG | GG | CC | CG | GG | |||||
| 295 (66) | 133 (29.8) | 19 (4.3) | 618 (64.3) | 304 (31.6) | 39 (4.1) | CD | 1.06 (0.79–1.41) (GG) | 0.71 | Cao 2013 | ||
| Dominant | 1.05(0.83–1.33) (CG+GG) | 0.64 | |||||||||
| 167 (69) | 61 (25.2) | 14 (5.8) | 183 (62.7) | 99 (33.9) | 10 (3.4) | CD | 0.68 (0.46–1.01) (CG) | 0.05 | Yang 2012 | ||
| Dominant | 0.66 (0.45–0.97) (CG+GG) | 0.03 | |||||||||
| rs36012910 | AA | AG | GG | AA | AG | GG | |||||
| 288 (84.71) | 51 (15) | 1 (0.29) | 234 (93.23) | 17 (6.77) | 0 | CD | 2.44 (1.37–4.33) (AG) | 0.002 | Wu 2012 | ||
| Dominant | 2.36 (1.35–4.14) (AG+GG) | 0.002 | |||||||||
| rs11887120 | TT | TC | CC | TT | TC | CC | |||||
| 57 (23.6) | 121 (50) | 64 (26.4) | 74 (25.3) | 155 (53.1) | 63 (21.6) | CD | 1.37 (0.88–2.13) (AA) | 0.17 | Wang 2015 | ||
| rs13428812 | AA | AG | GG | AA | AG | GG | |||||
| 137 (56.6) | 84 (34.7) | 21 (8.7) | 160 (55.4) | 106 (36.7) | 23 (8) | CD | 1.11 (0.58–2.12) (GG) | 0.84 | Yang 2012 | ||
| Dominant | 0.91 (0.64–1.31) (AG+GG) | 0.63 | |||||||||
Genotyping results for DNMT3A of all studies included.
| Gene | SNP | Genotype distribution n (%) | Analysis model | OR (CI) | p value | Reference | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Cases | Controls | ||||||||||
| DNMT3B | rs6119954 | GG | GA | AA | GG | GA | AA | ||||
| 200 (44.8) | 199 (44.6) | 4 (10.6) | 433 (45.1) | 445 (46.3) | 83 (8.6) | CD | 1.37 (0.88–2.13) (AA) | 0.17 | Wang 2015 | ||
| rs1569686 | TT | TG | GG | TT | TG | GG | |||||
| 360 (80.5) | 82 (18.3) | 5 (1.1) | 801 (83.4) | 150 (15.6) | 10 (1) | CD | 1.33 (0.41–4.31) (GG) | 0.64 | Wang 2015 | ||
| 230 (88.8) | 27 (10.43) | 2 (0.77) | 203 (77.48) | 55 (20.99) | 4 (1.53) | Dominant | 0.43 (0.26–0.72) (TG+GG) | <0.01 | Hu 2010 | ||
| rs4911107 | AA | AG | GG | AA | AG | GG | |||||
| 360 (80.5) | 82 (18.3) | 5 (1.1) | 800 (83.2) | 151 (15.7) | 10 (1.1) | CD | 1.08 (0.13–8.86) (GG) | 0.94 | Wang 2015 | ||
| rs4911259 | GG | GT | TT | GG | GT | TT | |||||
| 359 (80.3) | 83 (18.6) | 5 (1.1) | 799 (83.1) | 152 (15.8) | 10 (1.1) | CD | 1.09(0.13–8.88) (TT) | 0.94 | Wang 2015 | ||
| rs8118663 | AA | AG | GG | AA | AG | GG | |||||
| 126 (28.2) | 233 (52.1) | 88 (19.7) | 314 (32.7) | 473 (49.2) | 174 (18.1) | CD | 1.32 (0.91–1.91) (GG) | 0.15 | Wang 2015 | ||
| rs2424908 | TT | TC | CC | TT | TC | CC | |||||
| 78 (32.2) | 114 (47.1) | 50 (20.7) | 99 (33.7) | 139 (47.3) | 56 (19.1) | CD | 1.05 (0.64–1.71) (CC) | 0.96 | Yang 2012 | ||
| Dominant | 0.96 (0.68–1.36) (TC+CC) | 0.82 | |||||||||
| rs2424913 | CC | CT | TT | CC | CT | TT | |||||
| 257 (99.2) | 2 (0.8) | – | 259 (98.8) | 3 (1.2) | – | CD | 1.49 (0.17–17.94) (CT) | >0.05 | Hu 2010 | ||
| 152 (100) | – | – | 247 (100) | – | – | NA | NA | NA | Aung 2005 | ||
| 205 (96.7) | 7 (3.3) | – | 279 (94.9) | 15 (5.1) | – | Dominant | 0.66 (0.26–1.67) (CT) | 0.38 | Wang 2005 | ||
NA, not available; CD, co-dominant; NR, no risk; DNMT1 – DNA (cytosine-5)-methyltransferase 1; DNMT2 – TRNA (cytosine38-C5)-methyltransferase; DNMT3a – DNA (cytosine-5)-methyltransferase 3 alpha; DNMT3b – DNA (cytosine-5-)-methyltransferase 3 beta; SNP – Single Nucleotide Polymorphism; Allele: A – Adenine, C – Cytosine, G – Guanine, T – Thymine; OR (95% CI) – Odds Ratio (95% Confidence Interval).
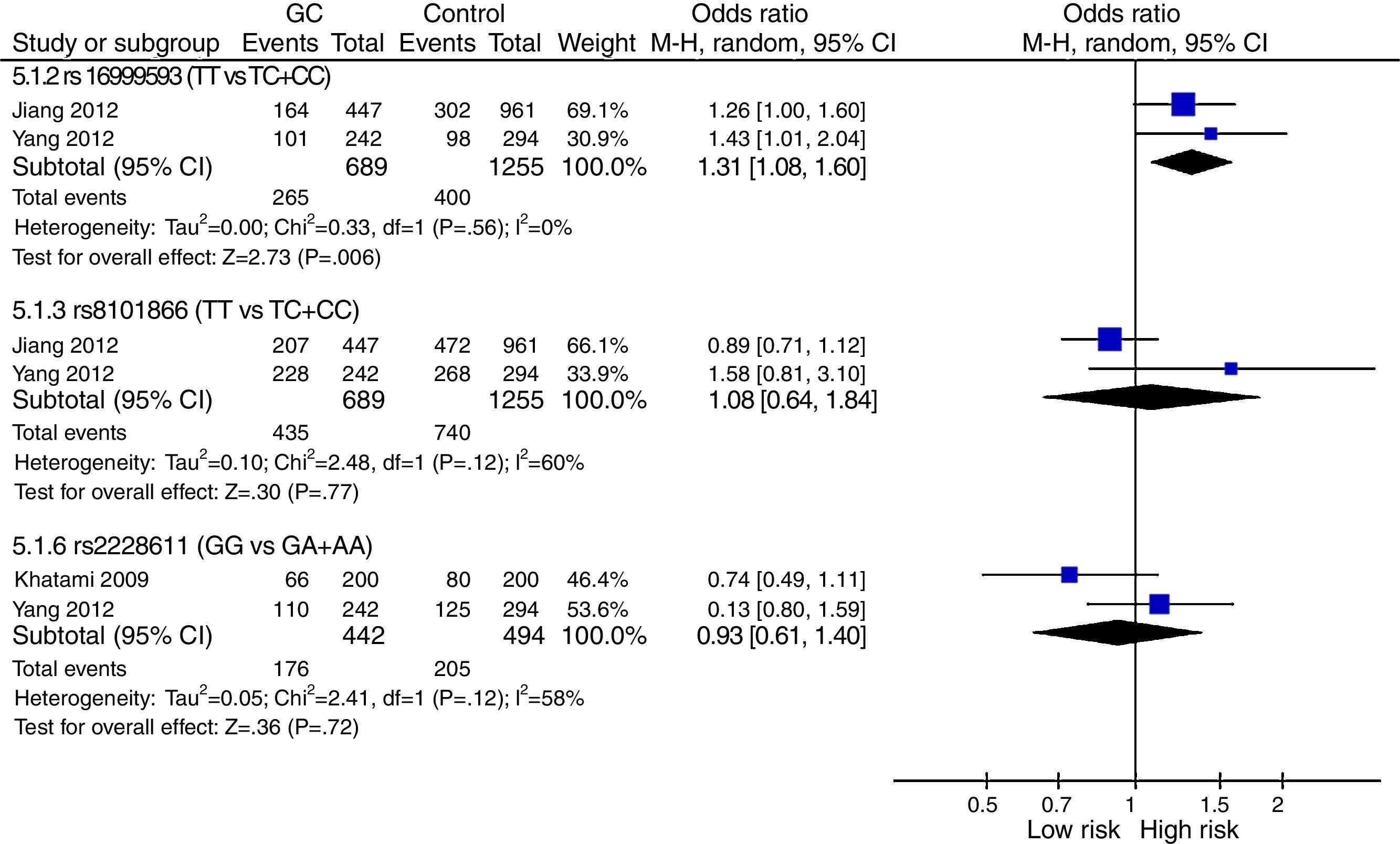
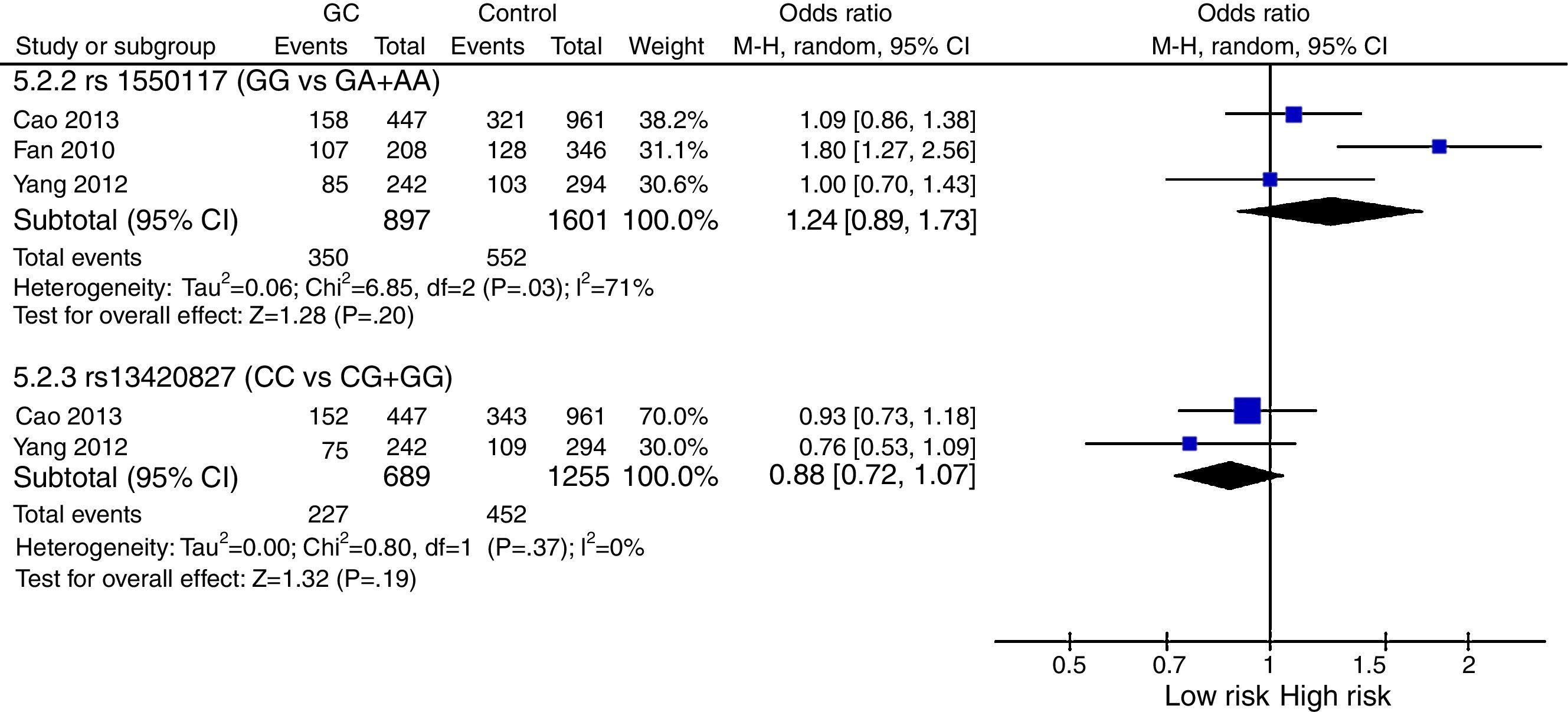
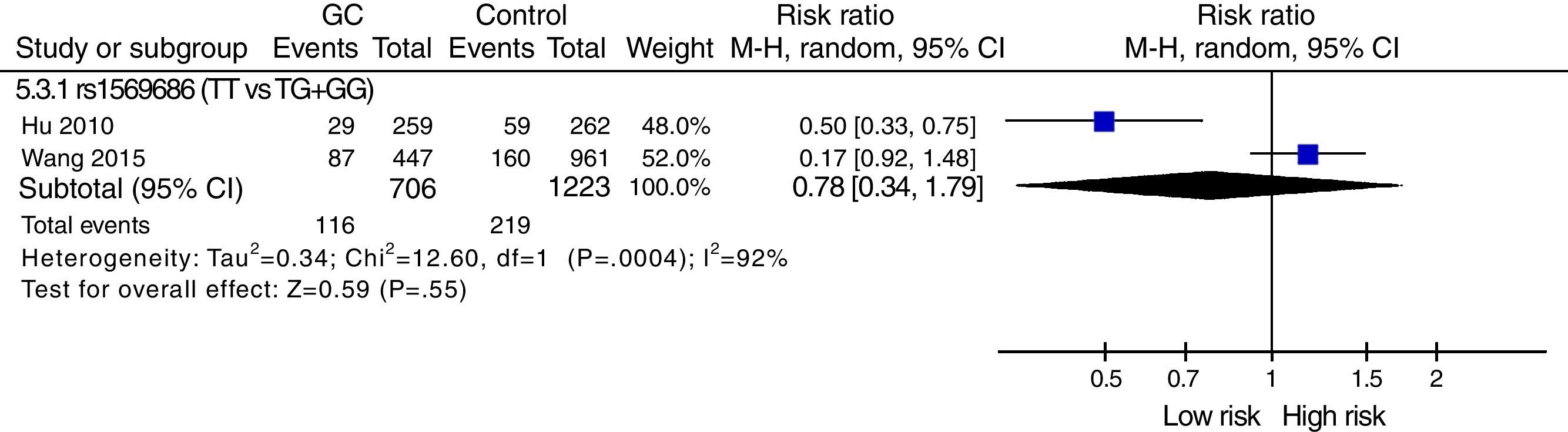
The meta-analysis was performed with data from all SNPs evaluated in more than one study – Figs. 2–4. A total of 6 SNPs were analyzed: DNMT1 rs16999593, rs8101866 and rs2228611; DNMT3A rs1550117 and rs13420827; and DNMT3B rs1569686. For the analysis, we have used the dominant genetic model, considering the risk genotype as the presence of any copy of the less frequent allele. We have also tested all data considering the Recessive Model for each SNP, and no significant change was observed in the reported data (data not shown). The majority of studies showed no significant heterogeneity in the data reported (p>0.05).
DNMT1: The analysis revealed no genetic heterogeneity (p>0.05) amongst the three SNPs tested. We found that only rs16999593 had a statistically significant result, revealing a 31% increased risk of GC development for TC+CC genotypes (p=0.006; OR=1.31; 95%CI=1.08–1.60). The rs8101866 and rs2228011 showed no significant differences (p>0.05) and no impact on the risk (OR=1.08 and OR=0.93, respectively) – Fig. 2.
DNMT3A: Statistical analysis showed heterogeneity of studies for rs1550117 (p=0.03), and moreover no impact on the risk of GC development for either rs1550117 or rs13420827 (p=0.20 and p=0.19, respectively) – Fig. 3.
DNMT3B: One SNP (rs2424913) was excluded from the meta-analysis because there was no data for all genotypes in the three studies, making it impossible to estimate the OR value.9,23,24 The analysis also revealed that studies regarding rs1569686 are very contradictory, resulting in a high heterogeneity on studies (p<0.001) and no association with GC – Fig. 4.
DiscussionGastric carcinogenesis is a multistep process, where different factors are involved and epigenetic alterations seem to play an important role in early mechanisms of this process.6 DNA CpG methylation is catalyzed by DNMTs and modified DNA methylation has direct effect on the regulation of gene expression. Considering the significant impact of genetic susceptibility based on the genetic sequences of individuals, functional SNPs may affect DNMTs expression, and therefore, it is important to study SNPs in DNMTs and understand the effect that functional SNPs can have on protein activity.
Literature reveals that DNMTs are frequently overexpressed in GC.14 Recently, several studies have been performed to evaluate the impact of DNMTs polymorphisms in GC susceptibility.3,8–11,19–24 Hence, this study intends to resume the data published regarding the association of SNPs in DNMTs with GC. By searching literature, we have found 11 studies matching our inclusion criteria. One of the first findings was that the majority of included studies were performed in Asiatic populations, where GC is extremely common, and therefore this is a major limitation since that is not possible to extrapolate the conclusions to others populations.25
A total of 24 SNPs were found in this systematic review: 10 in DNMT1, 1 in DNMT2 and 13 in DNMT3. The heterogeneity found among the included studies is due not only to population differences but also to different genotyping methods. The genotyping methods used had different sensitivity/specificity: PCR-RFLP method has some limitations to genotype correctly the three genotypes, specially heterozygote genotypes26; while TaqMan real-time PCR assays are more accurate and have less limitations and are a better candidate for SNP genotyping.27 Furthermore, the number of cases and controls included are extremely different among studies and therefore more studies with bigger populations and more precise genotyping methods are still required.
Our results revealed that DNMT1 rs16999593, DNMT2 rs11254413 and DNMT3A rs7560488 and rs36012910 are the only SNPs associated with GC.3,8,19 The DNMT1 rs16999593 is characterized by a C>T variation, resulting in an Arg to His amino acid substitution at position 97 of the protein, leading to missense mutation that may affect the structure and function of DNMT1. Literature demonstrated that rs16999593 is significantly associated to different malignancies: Tao et al. showed that individuals with TC genotype had a 4-fold increased risk for sporadic triple-negative breast carcinoma28; Xiang et al. also revealed that rs16999593 can provide protective effect for ductal breast carcinoma29; and Yang et al. reported a 1.45-fold increased risk for TC+CC carriers and GC.3 Although these evidences, other studies do not show any association.11,30 DNMT2 rs11254413 is characterized by a G>A substitution leading to an amino acid change, His to Tyr at position 101 of the protein that may affect DNMT2 function.3 The A-allele was associated with protection risk for GC by Yang et al., nevertheless, there is no functional study that support the biological role of this SNP on DNMT2 function.3 The DNMT3A rs7560488 (variation T>C) is a tagSNP that represents SNPs of the DNMT3A1 promoter with high linkage disequilibrium and has been associated with higher risk for GC in Asiatic population.8 Authors suggested that T to C change influences the binding of transcriptional factors and consequently expression levels of DNMT3A.8 The DNMT3A rs36012910, which is characterized by a A>G substitution, is located in the promoter region of DNMT3A and the substitution has been associated with DNMT3A altered activity.19 Wu et al. reveals that AG+GG genotypes were associated with a higher risk for GC.19
Regarding the data from the meta-analysis, we only were able to compare the results of 6 SNPs, which were described in more than one study. For DNMT1 analysis, 3 SNPs were studied (rs16999593, rs8101866 and rs2228611) and only rs16999593 showed statistically significance3,22 – Fig. 2. The results showed an increased risk for TC+CC genotypes of GC development. Nevertheless, the low number of studies and the lack of studies from populations apart from China make it necessary to perform more studies with different populations to confirm the association of rs16999593 with GC. Concerning DNMT3A, 2 SNPs were included in the meta-analysis (rs1550117 and rs13420827) but no association with GC was found – Fig. 3. The analysis revealed heterogeneity in rs1550117 distribution in populations, especially when comparing the report by Fan et al. to the other studies (Fan et al., 2010). Nevertheless, even removing Fan et al. from the analysis there was no significant association of rs1550117 with GC development. Furthermore, the differences in genotype distribution might be explained because of different genotype methods used in the three studies – Table 1. Finally, we included one SNP in DNMT3B on the meta-analysis (rs1569686) that showed no association with GC – Fig. 4. The analysis revealed heterogeneity in the distribution and a significant difference in the OR of the two studies (OR=0.43 in Hu et al. and OR=1.21 in Wang et al.).9,10
ConclusionsWe have found 11 articles studying SNPs in DNMTs but the variety of SNPs and the reduced number of studies per each SNP made this analysis complicated, allowing us to draw only some remarks and not strong conclusions. The data from studies revealed that rs11254413, rs7560488, rs36012910 and, specially, rs16999593 seem to be good candidates to be studied for their role in GC development. Nevertheless, our meta-analysis revealed that only one SNP (rs16999593) in DNMT1 was associated with GC development, especially TC+CC carriers.
More studies with different populations are required to prove if the SNPs selected in our analysis are associated with GC and furthermore, studies regarding biological importance of functional SNPs in DNMTs activity and expression are also needed to fully understand the impact of these SNPs in DNMTs on GC.
Conflicts of interestThe authors declare no conflicts of interest.