Potato common scab is caused by several soil-inhabiting pathogenic Streptomyces species. In the present study, a species-specific PCR method was used to detect Streptomyces species in potato tuber lesions and soils. Total genomic DNA from soil samples from six locations and tuber samples from four potato cultivars (Spunta, Shepody, Innovator and Russet Burbank) were assessed. Streptomyces scabies, Streptomyces acidiscabies, and Streptomyces turgidiscabies were detected in soybean, tobacco and potato soils and in all potato varieties except Russet Burbank. The phylogenetic analysis of the sequences obtained confirmed the identification. The method proposed proved to be time-saving and cost effective for the rapid detection of Streptomyces species. This is the first report of the detection of S. acidiscabies and S. turgidiscabies in soils and potato tubers from Argentina.
La sarna común de la papa es causada por varias especies patogénicas de Streptomyces habitantes del suelo. En el presente estudio se utilizó un método basado en PCR especie-específico para detectar las especies de Streptomyces presentes en lesiones de tubérculos de papa y suelos. Se extrajo ADN genómico total de muestras de suelo de seis localidades y cuatro cultivares de papa (Spunta, Shepody, Innovator y Russet Burbank).Streptomyces scabies, Streptomyces acidiscabies y Streptomyces turgidiscabies fueron detectadas en suelos cultivados con soja, tabaco y papa, y además, en todas las variedades de papa excepto las de Russet Burbank. El análisis filogenético de las secuencias confirmó las identificaciones. Se comprobó que el método propuesto brinda una rápida detección de las especies de Streptomyces. Este es el primer informe sobre la detección de S. acidiscabies y S. turgidiscabies en suelos y tubérculos de papa de Argentina.
In Argentina, annual potato production is almost 2 million tons3, and scab disease significantly damages tubers and other root crops. Streptomyces scabies, Streptomyces acidiscabies, and Streptomyces turgisdiscabies are the bestknown causal agents of this disease12. S. scabies occurs worldwide; S. acidiscabies was isolated in the United States, Japan, and Korea, while S. turgidiscabies was isolated in the United States, Japan, and Finland15. In Argentina, S. scabies was identified in 1935 from potatoes with scab symptoms and later isolated from 53 locations in 12 provinces, showing the widespread distribution of the pathogen in soils9.
The characteristics and pH of the soil can greatly affect the severity of potato scab. Scab is most severe in soils with pH values from 5.2 to 7.0. In some cases, scab control can be achieved by lowering the soil pH. However, “acid scab” is caused by S. acidiscabies in soils with pH of 5.0 or below6. Thus, measuring the soil pH may be useful to predict the pathogen attack to susceptible potato cultivars if the presence of the pathogen is detected in the soil.
Isolation and identification of Streptomyces from potato tubers and soil is time-consuming and laborious owing to the slow growth rate of bacteria and the high diversity of Streptomyces species inhabiting scar lesions and soil5.
The polymerase chain reaction (PCR) assay is less timeconsuming and superior to other techniques in its simplicity, rapidity, and sensitivity12. Kageyama et al.4 developed a PCR protocol for detection of plant fungal pathogens in the soil. The combination of this method with species-specific primers to amplify the genes of Streptomyces spp. could be a useful tool to detect the pathogen in the environment. In the last decade, Tanaka14 designed species-specific primer combinations to amplify part of the 16S rRNA and Internal Transcribed Spacer (ITS) genes of S. scabies, S. acidiscabies, and S. turgisdiscabies to detect Streptomyces species pathogenic on potato tubers. Thus far, in Argentina Streptomyces species have been identified in soils by morphological analysis of the isolates. This study proposes a diagnostic test that does not depend on the isolation, but allows Streptomyces species to be detected in soils in a faster and more accurate way, complementing the traditional method. Therefore, the goal of the present research was to detect Streptomyces species associated with potato common scab in Argentina using a speciesspecifi c PCR method.
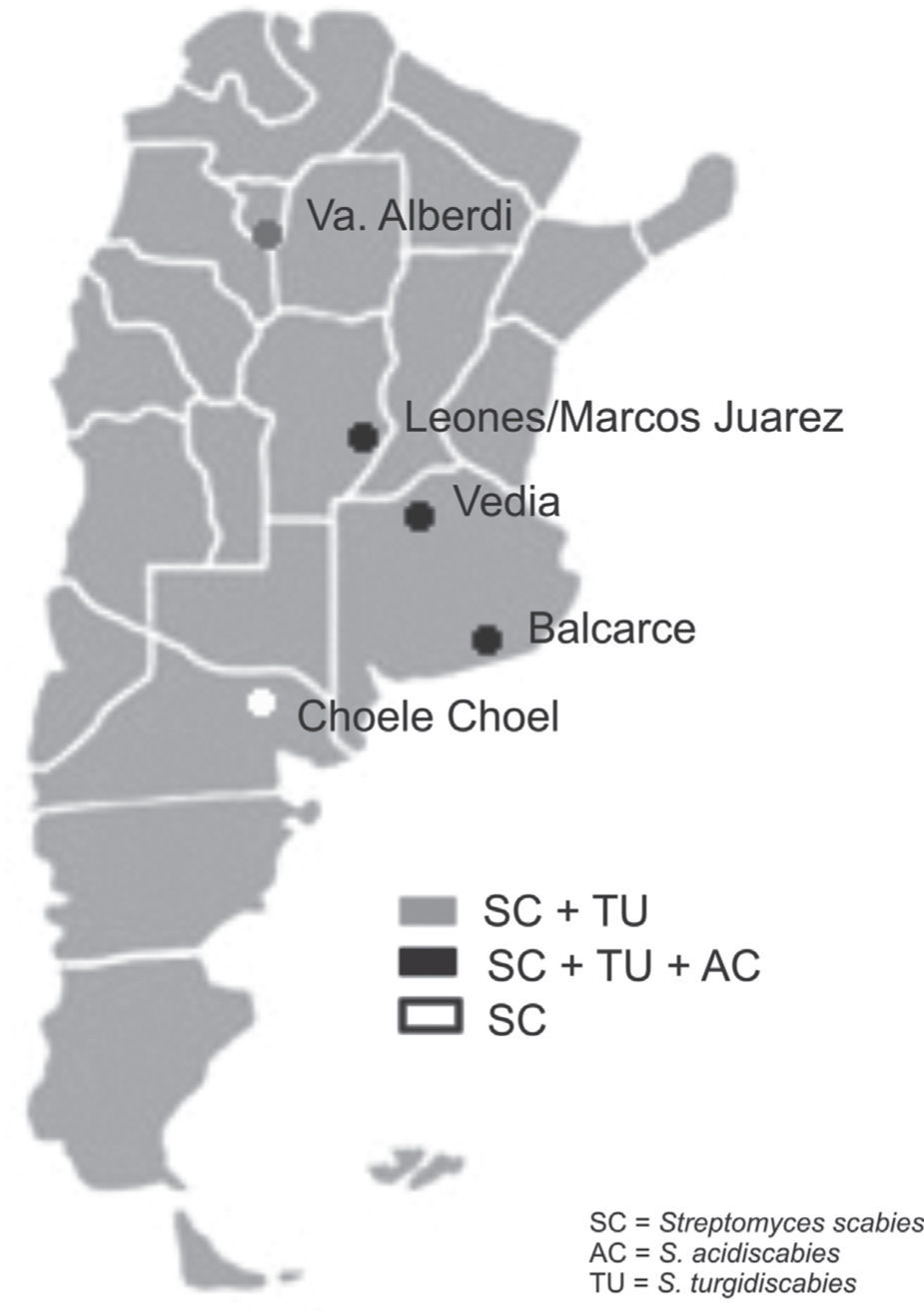
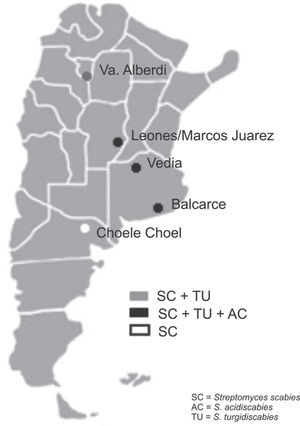
Soil samples having different soil textures and crop history were collected from six locations in four provinces of Argentina: 1) Balcarce (37°45'S, 58°18’W), Buenos Aires province, silty loam soil (Typic Argiudoll), field with long potato crop history; 2) Vedia, (34°29’W, 61°32’W), Buenos Aires province, sandy loam soil (Entic Hapludoll), soybean crop; 3) Marcos Juárez (32°41'S,62°09’W), Córdoba province, loam soil (Typic Argiudoll), soybean crop; 4) Leones (32°67′S, 62°30′W), Córdoba province, loam soil (Typic Argiudoll), soybean crop; 5) Choele Choel (39°16′S, 65°40′W), Río Negro province, sandy loam soil (Typic Natrargides), field with recent potato crop history; 6) Juan Bautista Alberdi (27°34′S, 65°37′W), Tucumán province, loam soil (Typic Argiudoll), tobacco crop. The soil was classified according to the USDA Soil Taxonomy.
The pH values of the soil samples were measured in filtrates of soil (100g/l) suspended in sterilized distilled water. The soil suspension was shaken in an orbital shaker at 200 r.p.m. (0.5% excentricity), 29°C±0.5°C for 90min, and pH measured with a pH meter (SANXIN, Shangai). This operation was repeated three times for each soil sample.
Potato tissue samples were collected from harvested tubers with scab symptoms from Balcarce, a location with more than 110 years of potato crop history. Thirty-one scab lesions were sampled from the following potato cultivars: 1) six lesions from three tubers of the cultivar Spunta; 2) twenty lesions from six tubers of the cultivar Shepody; and 3) five lesions from one tuber of the cultivar Innovator. Potato tubers of the cultivar Russet Burbank from Choele Choel, a location with only two years of potato crop history, were also evaluated. Since these tubers showed no scab lesions, no tissue samples were taken.
Total genomic DNA was extracted from soils according to Kageyama et al.4 as follows: a mixture of 0.2g soil sample and 0.2g of sterilized glass beads (1mm diameter) was suspended in 250μl extraction buffer: 100mM Tris-HCl (pH 9.0), 40mM EDTA, 2% (w/v) sodium dodecyl sulfate (SDS), 0.8% (w/v) skim milk (Difco, USA), and RNase A 200μg/ml (Nippongene, Japan), vortexed for 1min. Benzyl chloride (Fluka, Switzerland) (150μl) was added to the tube, vigorously vortexed for 2min, and then incubated at 50°C for 1h. Following this incubation, 150μl of 3M NaOAc was added to the suspension, lightly vortexed, and the mixture was incubated on ice for 15min. This suspension was cleared by centrifugation at 18,000g for 10min, and the upper layer was transferred to a clean tube. This step was repeated twice. DNA was precipitated with an equal volume of isopropanol and collected by centrifugation at 18,000g for 20min. The resulting pellet was rinsed with 70% ethanol and dried under vacuum. The DNA pellet was dissolved in 200μl of TE buffer [10mM Tris-HCl (pH 7.5), 1mM EDTA]. Three replicates were evaluated from each soil sample. Tissue samples from surface-washed potatoes were cut with a sterilized scalpel and chopped finely, before applying the same protocol as that applied to soil samples. PCR reactionswere performed with three different sets for each primerpair, in a total volume of 50μl reaction mixture containing 1μM of each primer, 1.25 units of Fast Start Taq DNA polymerase (Roche), 0.2mM dNTP mixture, 1×PCR buffer (10mM Tris/HCl, pH 8.3, 50mM KCl and 1.5mM MgCl2), 400ng/μl albumin from bovine serum (WAKO, Japan) and 1μl of DNA template. Three species-specific primer sets designed by Tanaka14 that partially amplify the 16S rRNA gene and the Internal Transcribed Spacer (ITS) was used in different amplification reactions. The primer sequences were as follows: AC-03 (5′-TACCGGATATCACTCCTGCCT-3′) and AC- 04 (5′-GGCGACCATCTCTGGCCGTT-3′) for S. acidiscabies (AC), SC-05 (5′-CCGGTAGCCCAACCCGTAAG-3′) and SC-06 (5′-GTAGTACTCACAGCCTCCGG-3′) for S. scabies (SC), and TU-01 (5′-GGAAACATCCAGAGATGGGTG-3′) and TU-02 (5′-GACAGTACTGGAAGGAGAAGA-3′) for S. turgidiscabies (TU). The molecular sizes of the PCR products were 836 bp, 236 bp, and 686 bp, respectively. The universal primers FM58 (5′-CCACAAATTTCACTACATTGA-3′) and FM66 (5′-TAGGATTTCAAGATCCTGC-3′) for the amplification of mitochondrial cytochrome oxidase II (coxII gene), which yielded the 600 bp expected band, were used as positive control8 to confirm the success of the DNA extraction. The reactions were carried out with a Mastercycler® gradient thermocycler (Eppendorf, Germany). The temperature cycling parameters were programmed for one cycle of 5min at 95°C, followed by 40 cycles of 30 sec at 94°C, 30 sec at 60°C (FM and TU) and 64°C (SC and AC) of annealing step, 2min at 72°C, and one cycle of 10min at 72°C. PCR products were electrophoresed in a 2% agarose LO3 (Takara, Japan) gel in TAE buffer (40mM Tris/HCl, pH 7.5, 19mM glacial acetic acid, and 2mM EDTA) and stained with ethidium bromide.
PCR products were extracted from the agarose gel using the Get pure DNA-Agarose kit (Dojindo Lab., Japan) following the manufacturer's instructions. The Big Dye™ Terminator Cycle Sequencing Ready Reaction Kit (Applied Biosystems, USA) was used for the sequence reaction according to the manufacturer's instructions. Electrophoresis of the sequencing products was carried out with an ABI 3700 DNA sequencer (Applied Biosystems). A phylogenetic analysis was performed to ensure the amplified fragments corresponded to the 16S rRNA gene and ITS. Sequences from GenBank corresponding to different Streptomyces species and the corresponding sequences obtained from samples were aligned with the ClustalW algorithm. Streptomyces albidochromogenes and Streptomyces setonii were applied as outgroups. The evolutionary history was inferred using the Maximum Parsimony (MP) method using MEGA version 513. The Bootstrap analysis with 1,000 replicates was performed. The MP tree was obtained using the Close- Neighbour-Interchange algorithm with search level 3, and the initial tree was obtained with random addition of sequences. All positions containing gaps and missing data were eliminated from the dataset.
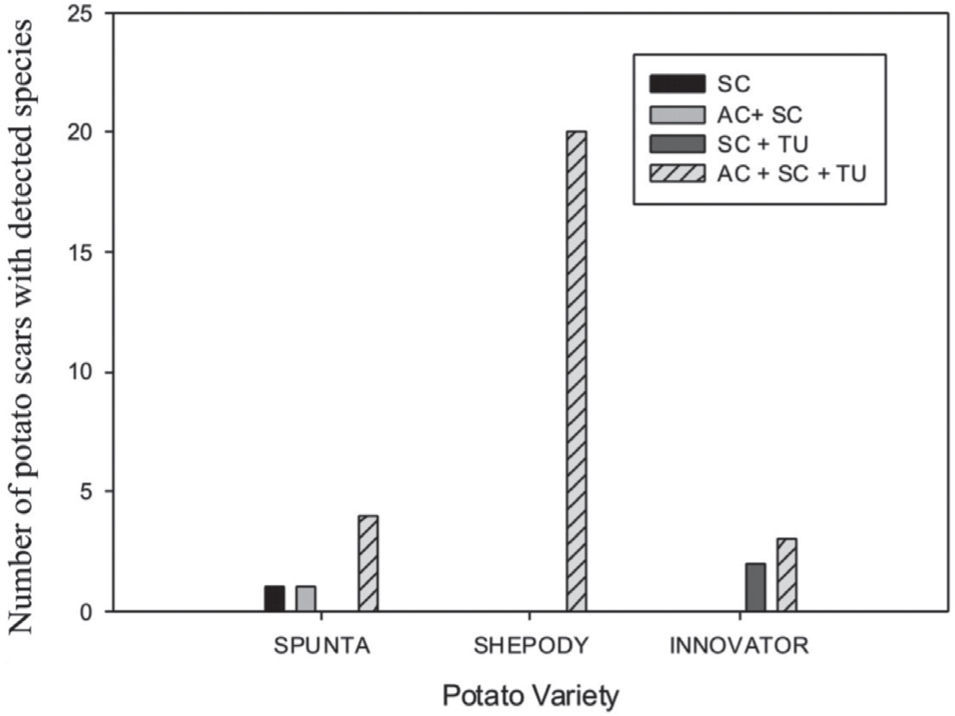
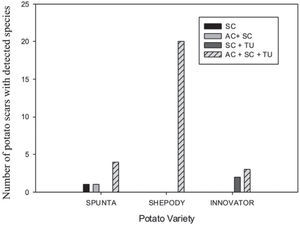
A scale was made with the presence of the species detected on the tuber lesions, either alone or in combination. The scale was as follows: 1: AC, 2: SC, 3: TU, 4: AC+SC, 5: AC+TU, 6: SC+TU, 7: AC+SC+TU. A contingency frequency distribution table was constructed with the pathogen and the cultivar as variables. The Pearson Chisquare was calculated to estimate the significance with α=0.5. A histogram with the combined variables was constructed.
The three Streptomyces species were detected in the three soybean fields and in the one with a long potato crop history. S. scabies was detected in all six soil samples. S. acidiscabies was not detected in Juan Bautista Alberdi or in Choele Choel, whereas S. turgidiscabies was not detected in Choele Choel (fig. 1). Non-specific bands amplified for S. scabies and S. acidiscabies, but they were smaller than the specific bands and could be avoided by increasing the annealing temperature. The pH values of the soils ranged from 6.1 to 6.5 (data not shown), Choele Choel being the least acid soil (pH=6.5).
The 31 scab lesions evaluated showed that with the simultaneous occurrence of the three species, the frequency was higher (87%) than with the occurrence of one or two species, in which cases frequencies were of 10% and 3%, respectively. These frequencies showed significant differences in the Pearson Chi-square analysis (χ2=19.82; 6 d.f.; p=0.003) among the Streptomyces species detected by scar and also among the potato cultivars (fig. 2).
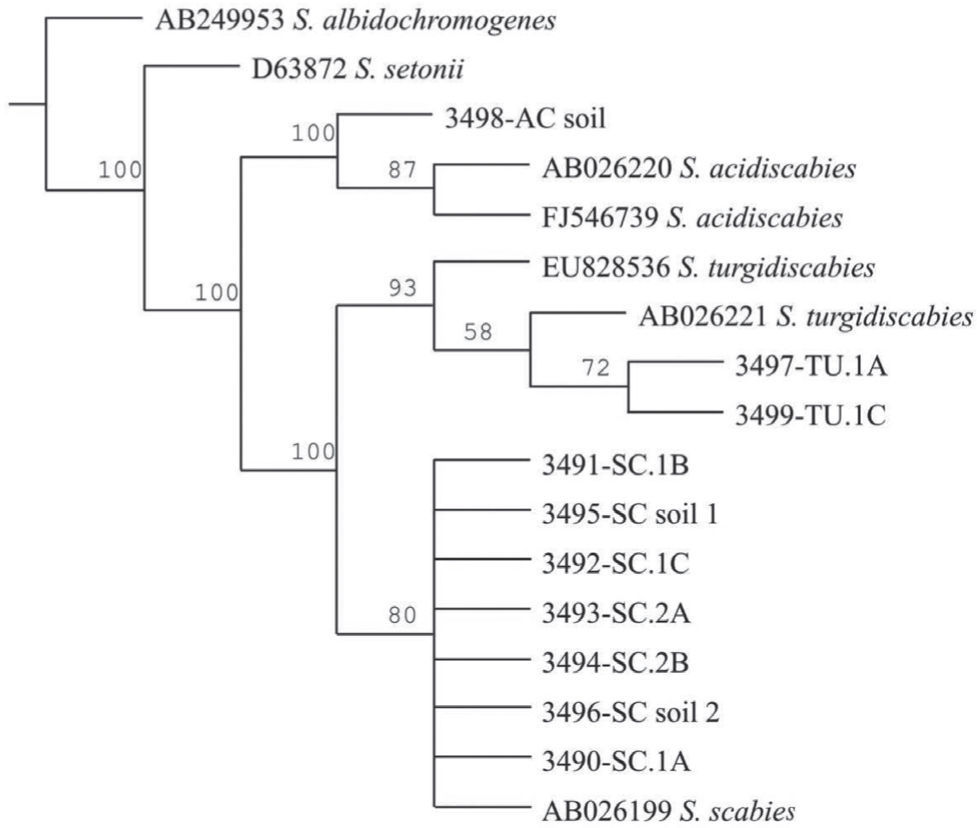
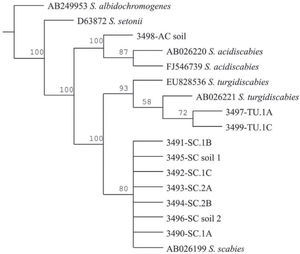
The Maximum Parsimony phylogenetic tree confirmed that the PCR products amplified by the species-specific primers corresponded to the reference sequences. The sequences analyzed formed two main clades: onecorresponding to S. acidiscabies supported by a bootstrap value (bv) of 100%, the other one grouping the species S. scabies and S. turgidiscabies supported by 100% bv. The subclade with S. turgidiscabies was supported by 93% bv, and the S. scabies group was supported by 80% bv (fig. 3).
Strict consensus phylogram of Maximum Parsimony Analysis of partial 16S rRNA and ITS sequences from Streptomyces taxa. The tree shows the position of the genomic DNA from soils and tubers, 3,491 to 3,499. There were a total of 136 positions in the final dataset, out of which 106 were parsimony informative. Five optimal trees were obtained; 1,106 steps; consistency index: 96; retention index: 95. The numbers above branches represent the bootstrap values. S. albidochromogenes and S. setonii are outgroups. SC represents the diagnostic band corresponding to S. scabies, AC corresponds to S. acidiscabies, and TU corresponds to S. turgidiscabies.
Our results showed that the combination of the PCR method with the species-specific primers proved to be efficient at detecting S. scabies, S. acidiscabies, and S. turgidiscabies in potato tuber lesions and in soils with different textures from Argentina.
Compared with previous studies using the plate cultivation method, the combined PCR method has notable advantages. Through this method, we were able to detect S. turgidiscabies and S. acidiscabies for the first time in Argentina. These findings were reported to SINAVIMO (Sistema Nacional Argentino de Vigilancia y Monitoreo de Plagas) as directed by SENASA (Servicio Nacional de Sanidad y Calidad Agroalimentaria). Furthermore, it was also possible to detect the three potato scab pathogens in scab lesion samples and soil samples.
The results of this study indicate that the three Streptomyces species may occur in soils with other crops, like soybean and tobacco. This result may be explained by the fact that all three species are known to be neither tissue- nor host-specific, enabling them to induce scab lesions on various root and tuber crops and to infect the fibrous roots of plants such as tobacco, soybean, and wheat1. According to Loria et al.7, the relevance of seedling infections is unknown, but the importance to pathogen survival should not be overlooked in studies of cropping systems targeted at scab management. In Balcarce, a location with a potato crop history of more than 100 years, all species were detected in the soil samples as well as in tubers of different cultivars, while the exclusive detection of S. scabies in soils in the Patagonian location of Choele Choel might stem from the fact that this is a relatively new potato production area under irrigation. The pH levels of the soils assessed would not limit the presence of the pathogenic Streptomyces species.
Cultivars Shepody, Spunta, Innovator, and Russet Burbank are recorded in Argentina as highly susceptible, susceptible, medium-resistant, and resistant to potato common scab, respectively11. In this study, the PCR detection method showed that the three Streptomyces species were detected in all the tubers of the cultivars Shepody, Spunta, and Innovator, with a higher frequency of the three species together in the same scab lesion. This feature supports the observations made by Park et al.10, Lethtonen et al.5, and Tagawa et al.12, who reported that multiple species may be present on scab lesions in infested tubers. The variable presence of Streptomyces species in different scars of the same tuber is an important issue because many results will depend on the selected number of scars per tuber for the isolation and identification of pathogenic Streptomyces species. Loria et al.7 concluded that the first step in developing scab control strategies for a production area should be a survey of the pathogenic species present in the environment, while Doroghazi et al.2 suggested that an understanding of the biogeography of Streptomyces should lead to the development of rational sampling strategies for discovering novel genetic diversity.
To our knowledge, this is the first report of the detection of S. acidiscabies and S. turgidiscabies in Argentina. It is clear that to extend the knowledge of the presence of these microorganisms in Argentina, the newly recorded species need to be isolated in future studies. In addition, an understanding of the biogeography of these organisms should lead to the development of rational cultural strategies for controlling them. The present method may be applicable as a complement to the culture methods, for quick detection of pathogenic Streptomyces in the soil, prior to crop management decisions.
Conflicts of interestThe authors declare that they have no conflicts of interest
We thank the Japan International Cooperation Agency (JICA) and the Instituto Nacional de Tecnología Agropecuaria (INTA) for funding this work. Special thanks to M.Sc. Dora Barreto for her contributions and to Lic. María José Beribe for the support with the statistical software.