The molecular basis of fluconazole resistance in Cryptococcus neoformans has been poorly studied. A common azole resistance mechanism in Candida species is the acquisition of point mutations in the ERG11 gene encoding the enzyme lanosterol 14-α-demethylase, target of the azole class of drugs. In C. neoformans only two mutations were described in this gene. In order to evaluate other mutations that could be implicated in fluconazole resistance in C. neoformans we studied the genomic sequence of the ERG11 gene in 11 clinical isolates with minimal inhibitory concentration (MIC) values to fluconazole of ≥16μg/ml. The sequencing revealed the G1855A mutation in 3 isolates, resulting in the enzyme amino acid substitution G484S. These strains were isolated from two fluconazole-treated patients. This mutation would not intervene in the susceptibility to itraconazole and voriconazole.
Las bases moleculares de la resistencia al fluconazol en Cryptococcus neoformans han sido poco estudiadas. Un mecanismo de resistencia a los azoles en Candida albicans es la adquisición de mutaciones puntuales en el gen ERG11, que codifica la enzima lanosterol 14 α-demetilasa, blanco de las drogas azólicas. En C. neoformans solo 2 mutaciones en este gen han sido descriptas. Con el objetivo de estudiar otras mutaciones que podrían estar implicadas en la resistencia al fluconazol en C. neoformans, realizamos la secuenciación del gen ERG11 de 11 aislamientos clínicos con valores de concentración inhibitoria mínima (CIM) de fluconazol ≥16μg/ml. En 3 aislamientos, la secuenciación reveló la mutación G1855A, que da como resultado la sustitución aminoacídica G484S. Estos aislamientos fueron recuperados de 2 pacientes tratados con fluconazol. Esta mutación no intervendría en la sensibilidad al itraconazol y al voriconazol.
Cryptococcosis is a life-threatening infection caused by the encapsulated basidiomycetous yeast Cryptococcus neoformans that affects mainly immunocompromised patients, especially those suffering from AIDS4. The most common manifestation is cryptococcal meningitis, which is fatal unless treated. C. neoformans is found worldwide and is responsible for approximately one million cases/year which result in over 600000 deaths annually7.
Fluconazole (FLC), a triazole antifungal drug, is the drug of choice for consolidation and maintenance therapy due to its efficacy, excellent central nervous system penetration and minor toxic effects9.
Due to the use of FLC in long-term therapies, there is concern about the emergence of antifungal resistance in C. neoformans3. Several authors have associated in vitro resistance with treatment failure and infection relapse1,3.
The molecular basis of resistance to azole antifungals has been poorly studied in C. neoformans.
One resistance mechanism proposed is the duplication of chromosome 1 and consequently of two of its resident genes: ERG11, which encodes for the FLC target enzyme lanosterol 14-α-demethylase, and AFR1, which encodes for an ABC transporter14. It has been demonstrated that upregulation of the AFR1 gene is involved in the resistance to FLC in this yeast11.
A common FLC resistance mechanism in Candida species is the acquisition of point mutations in the ERG11 gene resulting in an altered target with reduced affinity for or inability to bind azoles8. Only two mutations in this gene have been associated with resistance to FLC in C. neoformans10,13. Furthermore, one of them caused resistance to both FLC and voriconazole (VRC) and increased susceptibility to itraconazole (ITC) and posaconazole (PSC); this mutation was identified in an isolate with an exceptionally high level of heteroresistance13.
To elucidate if more mutations could be implicated in FLC resistance, we studied the ERG11 genomic sequence of eleven clinical isolates from the Mycology Department Culture Collection (DMic) of Instituto Nacional de Enfermedades Infecciosas “Dr. Carlos G. Malbrán”, Buenos Aires, Argentina. The research proposal does not involve experimentation on humans and non-clinical samples were used. Including yeast isolates are anonymous and belong to the Mycology Department Culture Collection. These isolates were selected for having high minimal inhibitory concentration (MIC) values to FLC (MIC values ≥16μg/ml). One isolate with a lower MIC value was incorporated because it came from a patient who had presented an isolate with a high MIC value. The isolates and the patients’ clinical data are described in Table 1. All the isolates included in this study were C. neoformans var grubii genotype VNI determined by PCR-RFLP of the URA5 gene6.
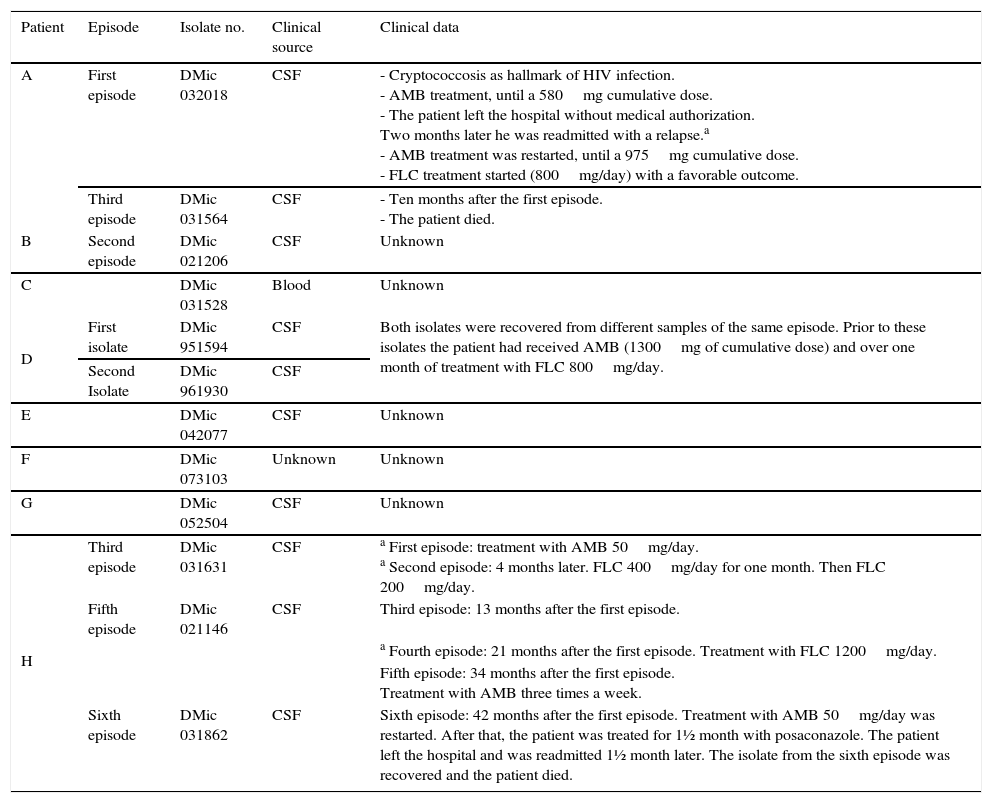
Medical records and clinical source of the Isolates
| Patient | Episode | Isolate no. | Clinical source | Clinical data |
|---|---|---|---|---|
| A | First episode | DMic 032018 | CSF | - Cryptococcosis as hallmark of HIV infection. - AMB treatment, until a 580mg cumulative dose. - The patient left the hospital without medical authorization. Two months later he was readmitted with a relapse.a - AMB treatment was restarted, until a 975mg cumulative dose. - FLC treatment started (800mg/day) with a favorable outcome. |
| Third episode | DMic 031564 | CSF | - Ten months after the first episode. - The patient died. | |
| B | Second episode | DMic 021206 | CSF | Unknown |
| C | DMic 031528 | Blood | Unknown | |
| D | First isolate | DMic 951594 | CSF | Both isolates were recovered from different samples of the same episode. Prior to these isolates the patient had received AMB (1300mg of cumulative dose) and over one month of treatment with FLC 800mg/day. |
| Second Isolate | DMic 961930 | CSF | ||
| E | DMic 042077 | CSF | Unknown | |
| F | DMic 073103 | Unknown | Unknown | |
| G | DMic 052504 | CSF | Unknown | |
| H | Third episode | DMic 031631 | CSF | a First episode: treatment with AMB 50mg/day. a Second episode: 4 months later. FLC 400mg/day for one month. Then FLC 200mg/day. |
| Fifth episode | DMic 021146 | CSF | Third episode: 13 months after the first episode. | |
| a Fourth episode: 21 months after the first episode. Treatment with FLC 1200mg/day. | ||||
| Fifth episode: 34 months after the first episode. Treatment with AMB three times a week. | ||||
| Sixth episode | DMic 031862 | CSF | Sixth episode: 42 months after the first episode. Treatment with AMB 50mg/day was restarted. After that, the patient was treated for 1½ month with posaconazole. The patient left the hospital and was readmitted 1½ month later. The isolate from the sixth episode was recovered and the patient died. |
CSF, cerebrospinal fluid; AMB, amphotericin B; FLC, fluconazole; PSC, Posaconazole.
The minimal inhibitory concentration (MIC) was determined according to the European Committee on Antimicrobial Susceptibility Testing (EUCAST) E.Def 7.2 reference document2. Amphotericin B (AMB) and ITC (Sigma–Aldrich Quimica, Argentina); FLC and VRC (Pfizer S.A., Argentina); were the drugs tested and were provided as standard powders of known potency. The susceptibility tests were repeated from 4 to 10 times for each isolate.
DNA was extracted according to the method reported by Möller et al. To obtain the complete sequence of the ERG11 gene, four PCRs were performed according to Rodero et al.10. PCR products were purified using the PureLink purification kit (Invitrogen) and were sequenced on both strands using the initial amplification primers with an automated DNA sequencer ABI Genetic Analyzer 3500 (Applied Biosystems, CA). Sequences were edited using the BioEdit Versión 7.0.0 (Tom May, North Carolina State University). All ERG11 gene sequences were deposited in the GenBank database (Table 2).
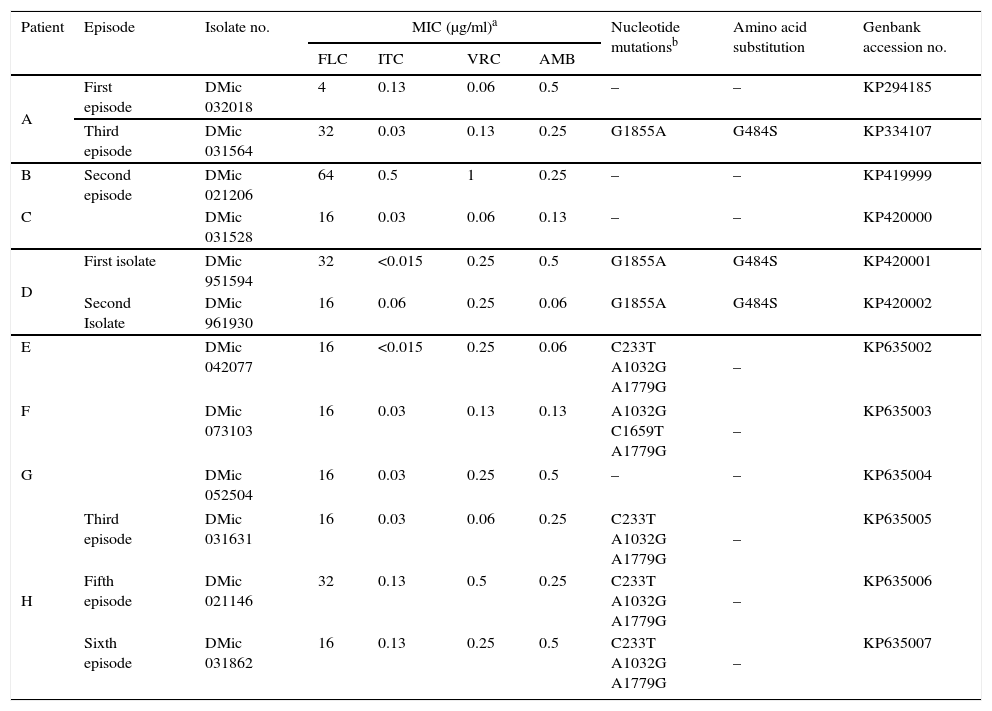
Antifungal susceptibilities, nucleotide mutations in the ERG11 gene and amino acid substitutions from Cryptococcus neoformans isolates
| Patient | Episode | Isolate no. | MIC (μg/ml)a | Nucleotide mutationsb | Amino acid substitution | Genbank accession no. | |||
|---|---|---|---|---|---|---|---|---|---|
| FLC | ITC | VRC | AMB | ||||||
| A | First episode | DMic 032018 | 4 | 0.13 | 0.06 | 0.5 | – | – | KP294185 |
| Third episode | DMic 031564 | 32 | 0.03 | 0.13 | 0.25 | G1855A | G484S | KP334107 | |
| B | Second episode | DMic 021206 | 64 | 0.5 | 1 | 0.25 | – | – | KP419999 |
| C | DMic 031528 | 16 | 0.03 | 0.06 | 0.13 | – | – | KP420000 | |
| D | First isolate | DMic 951594 | 32 | <0.015 | 0.25 | 0.5 | G1855A | G484S | KP420001 |
| Second Isolate | DMic 961930 | 16 | 0.06 | 0.25 | 0.06 | G1855A | G484S | KP420002 | |
| E | DMic 042077 | 16 | <0.015 | 0.25 | 0.06 | C233T A1032G A1779G | – | KP635002 | |
| F | DMic 073103 | 16 | 0.03 | 0.13 | 0.13 | A1032G C1659T A1779G | – | KP635003 | |
| G | DMic 052504 | 16 | 0.03 | 0.25 | 0.5 | – | – | KP635004 | |
| H | Third episode | DMic 031631 | 16 | 0.03 | 0.06 | 0.25 | C233T A1032G A1779G | – | KP635005 |
| Fifth episode | DMic 021146 | 32 | 0.13 | 0.5 | 0.25 | C233T A1032G A1779G | – | KP635006 | |
| Sixth episode | DMic 031862 | 16 | 0.13 | 0.25 | 0.5 | C233T A1032G A1779G | – | KP635007 | |
MIC, minimal inhibitory concentration; FLC, fluconazole; ITC, itraconazole; VRC, voriconazole; AMB, amphotericin B.
The FLC susceptibility testing confirmed that the strains selected for this study presented high MIC values (≥16μg/ml) (Table 2). All the isolates exhibited similar in vitro susceptibility patterns toward AMB, ITC and VRC as the FLC susceptible isolate included in this study and other isolates of our collection with FLC MIC values <16μg/ml (data not shown). Only one isolate (patient B, isolate no. DMic 021206) exhibited higher MIC values to ITC and VRC (0.5 and 1μg/ml respectively).
The Genbank accession numbers of the ERG11 gene sequences obtained are listed in Table 2. Eight of the eleven isolates studied contained nucleotide variations compared to the wild type published sequences of ERG11 (GenBank accession No. AY265353 and JQ044790). Only one of these nucleotide variations resulted in an amino acid substitution, the G1855A mutation producing substitution G484S. Two isolates recovered from the same episode of patient D and the isolate recovered from the third episode of patient A presented this mutation. We include in this study an isolate recovered from the initial episode of patient A, which did not contain this nucleotide variation and was susceptible to FLC. The other five isolates, obtained from three patients, contained different combinations of five nucleotide variations that did not result in any amino acid substitution: C233T present in an intron, and the silent nucleotide changes A1032G, C1659T, A1779G. Three isolates did not exhibit nucleotide variation compared to the published wild type sequence of the ERG11 gene.
The study of specific C. neoformans physiological responses and the possible resistance mechanisms to drugs used in the treatment of cryptococcosis are important both to identify potential new treatments for the infection and to enhance the inhibitory effects of existing drugs.
The ERG11 gene encodes the lanosterol 14-α-demethylase involved in ergosterol biosynthesis and the primary target for the azole class of antifungals. Several point mutations in this gene leading to different amino acid substitutions have been shown to decrease the target affinity for FLC resulting in drug resistance in C. albicans5,8. Two of them have been described and related to FLC resistance in C. neoformans: substitutions G484S and Y145F10,13.
In this study, three clinical isolates with high MIC values presented the G484S substitution. These isolates were recovered from two patients who had had previous cryptococcosis episodes with a history of treatment with FLC. Moreover, we were able to study the isolate obtained from one of these patients’ first episode, where cryptococcosis was the hallmark of HIV and the patient had not received any treatment. This initial isolate presented a lower MIC value and did not carry the amino acid substitution. These results reinforce the hypothesis that relates the G484S substitution to FLC resistance in C. neoformans. This relationship was proposed previously by Rodero et al. as a result of the study of a resistant isolate recovered from a patient suffering four episodes of relapse10.
According to the 3-dimensional model of Lanosterol 14-α-demethylase from C. neoformans, the amino acid G484 is located in the heme environment into the active site of the enzyme12. It is proposed that this amino acid substitution might decrease the flexibility required for binding with the substrate and the azole antifungal agents12.
Mutation G1885A leading to amino acid G484S substitution was found independently in isolates from different patients and may represent a “hot spot” for the development of azole resistance; furthermore this substitution correlates with substitution G464S in C. albicans also proposed as a “hot spot” for that species8.
The structure of VRC is very similar to FLC and in accordance with the three-dimensional models in C. neoformans, VRC might show higher affinity with the enzyme than FLC12. On the other hand ITC and PSC have very long side chains and might present the lowest interaction with the enzyme. With one exception, the isolates included in the present study exhibited low MIC values for VRC and ITC; moreover, we found no differences in the VRC and ITC MIC values between the isolates with the G484S substitution and others in our collection susceptible to FLC (data not shown), suggesting that the G484S substitution would not intervene in the enzyme interaction with ITC and VRC. In contrast, the other amino acid substitution, the Y145F, described in C. neoformans, afforded resistance to VRC but increased susceptibility to ITC and posaconazole13.
We also found different combinations of five nucleotide variations that did not result in any amino acid substitution, which may indicate allelic differences present in the ERG11 gene, and heterogeneity in the C. neoformans population. These allelic differences were also observed in C. albicans ERG11 gene8. It is worthy of note that all the isolates included in this study were C. neoformans var. grubii genotype VNI in line with the worldwide distribution since this genotype is the most ubiquitous and prevalent and causes most of the cryptococcal infections4,6.
Xu et al. proposed that mutation to FLC resistance in C. neoformans is a dynamic and heterogeneous process involving multiple simultaneous mechanisms15. Overexpression of efflux transporters and chromosome duplication may occur in the isolates without any amino acid substitution and may also be acting together with the G484S amino acid substitution. It remains to be determined how this mutation individually contributes to FLC resistance.
In summary, the results showed that FCZ resistance in C. neoformans may result from the presence of the G1855A point mutation in the ERG11 gene responsible for the amino acid substitution G484S. This mutation would not change susceptibility to ITC and VRC.
Ethical disclosuresProtection of human and animal subjectsThe authors declare that no experiments were performed on humans or animals for this study.
Confidentiality of dataThe authors declare that they have followed the protocols of their work center on the publication of patient data.
Right to privacy and informed consentThe authors declare that no patient data appear in this article.
Conflict of interestThe authors declare that they have no conflicts of interest.