Mine tailings contain high concentrations of heavy metals such as As, Pb, Cu, Mn, and Fe, which are detrimental to the health of humans and the environment. In tailings at the El Fraile mine in Guerrero, Mexico, some plant species are apparently tolerant of heavy metals and can be found growing in the tailings. These plants could be associating with heavy metal-tolerant bacteria that promote plant growth and improve biomass production, and these bacteria could be a useful alternative for bacteria-assisted phytoremediation. The objective of this study was to isolate bacteria detected in the mine tailings at El Fraile-Taxco, focusing on those in the soil from the rhizosphere, the inner tissue of the root, leachate, and water, which have the potential to promote plant growth. The ability of the isolated bacteria to promote plant growth was evaluated in vitro. Of the 151 morphotypes isolated, 51% fix nitrogen, 12% dissolve phosphates, and 12%, 39.7%, and 48.3% produce indole acetic acid, gibberellins, and siderophores, respectively. In addition, 66.7% were observed to produce lytic enzymes, such as proteases, celluloses, lipases, esterases, and amylases, which exhibited activity against Fusarium, Aspergillus, and Colletotrichum. The use of 16S rRNA analysis led to the identification of the bacterial genera Chryseobacterium, Bacillus, Pseudomonas, Mycobacterium, Staphylococcus, Curtobacterium, Enterobacter, Agrobacterium, Ochrobactrum, Serratia, Stenotrophomonas, and Acinetobacter. The bacteria isolated from the rhizosphere exhibited the greatest ability to fix nitrogen and produced indole acetic acid, gibberellins, siderophore, and lytic enzymes. In addition, the isolates collected from the soil samples demonstrated ability to solubilize phosphate.
Los jales mineros contienen una alta concentración de metales pesados como As, Pb, Cu, Mn y Fe. Estas altas concentraciones de metales son perjudiciales para la salud humana y el medio ambiente. En los jales mineros de El Fraile, México, es posible detectar especies de plantas tolerantes a los metales pesados; estas plantas podrían estar asociadas con bacterias capaces de promover su crecimiento, además de poseer actividad antagonista contra hongos. El objetivo de este estudio fue aislar de diferentes microambientes (suelo rizosférico, tejido de raíz, lixiviado y agua) del área del jale El Fraile bacterias con potencial de promover el crecimiento vegetal y actividad antagonista contra hongos fitopatógenos. Estudios in vitro demostraron que el 51% de los morfotipos aislados (151 en total) fijan nitrógeno y el 12% disuelven fosfatos. Asimismo, el 12, 39,7 y 48,3% producen ácido indolacético, giberelinas y sideróforos, respectivamente. Por otro lado, se observó que el 66,7% producía enzimas líticas como proteasas, celulasas, lipasas, esterasas y amilasas, además de exhibir actividad antagonista contra Fusarium, Aspergillus y Colletotrichum. Mediante análisis del gen 16S ARNr, se identificó a estas bacterias como pertenecientes a los géneros Chryseobacterium, Bacillus, Pseudomonas, Mycobacterium, Staphylococcus, Curtobacterium, Enterobacter, Agrobacterium, Ochrobactrum, Serratia, Stenotrophomonas y Acinetobacter. Las bacterias de la rizosfera exhibieron la mayor capacidad para fijar nitrógeno y produjeron ácido indolacético, giberelinas, sideróforos y enzimas líticas. Además, se detectó que las cepas aisladas de suelo rizosférico eran las que tenían la capacidad de solubilizar fosfatos.
The mine tailing deposits at El Fraile, located approximately 12km from the city of Taxco de Alarcon, cover an area of approximately 470m long and 372m wide, with a height of 60m, totalling approximately 5.5 million tons of material contaminated with potentially toxic elements (PTE) including Pb, Cd, Zn, Cr, Cu, V, Mn, Fe, and As42.
It has been shown that when mining residues cause heavy metals and metalloids to become mixed with the soil, the soil subsequently degrades. Mine drainage has been described to generate low-pH zones ranging from pH 2.0 to 5.0, which have been considered oligotrophic environments43. There are low levels of vegetation on mine drainage sites that vary depending on the conditions of the site and can include six types of vegetative communities, with tropical deciduous forests being the most abundant followed by conifers, mountain mesophyll, and semipermeable wetland grasslands3.
Thirteen plant species were isolated from the El Fraile mining zone13. The presence of this vegetation could be attributed to certain microorganisms with the ability to thrive in extremely contaminated environments. Such microorganisms can modify the chemical state, form or distribution of the heavy metals in the soil through redox processes49. Different bacterial genera have been isolated from mine tailings, soil, water, leachate, and wetlands. The strains that proved to be capable of surviving in these harsh conditions of heavy metals and metalloids include Pseudomonas, Staphylococcus, Enterobacter, and Chromobacterium46.
Detailed studies around the world have focused on the characterization of plant growth-promoting bacteria (PGPB). These bacteria are described as a heterogeneous group associated with the plant rhizosphere. This group of microorganisms improves plant growth by direct or indirect mechanisms. The primary characteristics of these bacteria include their ability to colonize the surface of roots and compete with other microorganisms by multiplying efficiently, while promoting plant growth and/or plant biocontrol2. Similarly, these bacteria possess the ability to transform the state of nutrients into forms that are accessible to plants through absorption by the roots. The bacterial genera reported to contain PGPB include Acinetobacter, Agrobacterium, Arthrobacter, Azoarcus, Azospirillum, Burkholderia, Enterobacter, Klebsiella, Pseudomonas, Serratia, Pantoea, and Thiobacillus, and other genera11,45. However, some species of Pseudomonas, Bacillus, Rhizobium, and Streptomyces have been reported to have stronger effects in many plants of agricultural interest when bacterial rhizosphere isolates are analyzed12. The growth promoting effects of PGPB may occur through direct mechanisms, such as biofertilisation, and/or indirect mechanisms, such as biocontrol26. Direct mechanisms include nitrogen fixation, phosphate solubilization, and auxin, gibberellin and ethylene production, respectively. Indirect mechanisms include the production of siderophores and lytic enzymes, such as proteases, elastases, esterases, and amylases6. Plant-bacteria associations can be utilized to improve biomass production and to rehabilitate soils contaminated with metals by employing technologies like phytoremediation and phytomining. Coupling traditional phytoremediation with PGPB is thus a very promising strategy for improving contaminated sites. If successful, this combined bioremediation process could increase the efficiency of rehabilitation and decrease exposure to potentially toxic elements (PTE), improving environmental and human health. Here, we evaluated the ability of bacteria isolated from the tailings at El Fraile to promote plant growth, with the intention of applying them to assist in phytoremediation in the near future.
Materials and methodsIsolation and selection of bacterial morphotypesThe El Fraile mine tailing site in Taxco de Alarcon, Guerrero, Mexico, has been studied for several years and the physicochemical properties of soil, leachate and water have been previously determined as follows. The pH of the soil is 2.3–2.9, and the heavy metals present in the soil are: Ag (9.5–74.2mg/kg), Cd (1.0–780mg/kg), Cu (71.8–1320mg/kg), Fe (2.49–25.1mg/kg), Mn (18.6–13800mg/kg), Pb (780–43700mg/kg), V (2.0–127mg/kg), Zn (380–>10000mg/kg) and As (19.0–11800mg/kg)42. Leachates have moderate to low pH (<5.5) and contain variable levels of SO4−2 (280–29500mg/l), As (<0.01–12.0mg/l), Fe (0.025–2352mg/l), Mn (0.1–732mg/l), Zn (<0.025–1465mg/l) and Pb (<0.01–0.351mg/l)43. The pH of the water is 6.9–7.1 and it contains SO4−2 (280–3024mg/l), Pb (<0.01mg/l), Fe (0.02–15.9mg/l), Mn (0.1–2.3mg/l), Zn (0.025–3.3mg/l), and As (0.01–0.3mg/l)43,44.
Samples were collected from within the El Fraile mining zone from five different substrates: soil, rhizosphere, root internal tissues, leachate, and water. Each sample consisted of 100g of material. Root samples and rhizosphere samples were taken from the plant Acacia farnesiana, which grows on the tailing piles and is reported to be a bioaccumulator of heavy metals3. The samples were transported in plastic bags, paper bags, and sterile flasks. Serial dilutions were made from 10−1 to 10−4 per 1g or 1ml of sample depending on its origin. One hundred microliters of the last dilution were used to inoculate LB agar. The plates were incubated at 30°C for 24–48h. Root tissue samples were cut and weighed to produce 1g samples and were then washed and disinfected under sterile conditions. The samples were transferred to 5ml of LB broth in tubes and incubated at 30°C for 24h. All isolated samples were serially diluted to 10−3, and 100μl was utilized as inoculum, as described for the previous samples. The isolates obtained from all of the samples processed were selected according to the morphological diversity of their colonies, and were replated to obtain pure isolates45.
In vitro screening of microorganism isolates for their plant growth-promoting (PGPB) activitiesNitrogen fixationTo evaluate the ability of the isolates to fix nitrogen from the atmosphere, they were inoculated by stabbing a modified nitrogen-free Rennie medium36. The isolates were then incubated at 30°C for 24–48h. Growth in this medium is indicative of nitrogen fixation. The positive control for this experiment utilized Klebsiella variicola T29A32.
Phosphate solubilizationThe ability to solubilize inorganic phosphate was evaluated in NBRIP media with tricalcium phosphate34. Isolates were inoculated via stabbing and incubated at 30°C for 96h. Clear halos around the colonies indicated that the isolates could solubilize inorganic phosphate. Each halo was measured to calculate the solubilization index.
Quantification of indole acetic acid (IAA)Indole acetic acid production was evaluated in trypticase soy broth supplemented with tryptophan. Isolates were inoculated and incubated for 96h at 30°C with shaking at 120rpm. To quantify the IAA, the cultures were centrifuged at 4000rpm for 20min, a 75-μl sample was removed from the supernatant and mixed with Salkowski reagent, and the reaction mixtures were left to incubate at room temperature in the dark. A positive result ranges from light pink to fuchsia. Absorbance was measured at 492nm in a Stat Fax-2100 reader27. To quantify the IAA produced by the isolates tested, a reference curve was generated with Salkowski reagent diluted 2:1 at different concentrations of commercial IAA (2, 4, 6, 8, 10, 15, 20, 30, 40, 50, and 60μg/ml) (I2886 Sigma-Aldrich). Azotobacter vinelandii was utilized as a positive control, and Salmonella Enteritidis was employed as a negative control for this experiment15.
Gibberellin detectionGibberellins were detected from cultures grown in trypticase soy broth supplemented with glucose. The isolates were inoculated and incubated for 96h at 30°C. Seventy-five microliters of the culture and 150μl of Folling-Wu reagent were transferred to a 96-well plate. Isolates that were positive for this test exhibited a green-blue coloration due to the reduction caused by molybdic acid in the presence of gibberellic acids14.
Siderophore detectionBacteria were inoculated into King's B broth containing the following ingredients; protease peptone (20g/l), dipotassium hydrogen phosphate (1.5g/l), magnesium sulphate heptahydrate (1.5g/l), and supplemented with FeCl3 (50mg/l). Incubation was performed at 30±1°C for 48h26. To visualize cultures capable of siderophore production, each culture was exposed to UV light in a mini transilluminator (BioRad) to observe fluorescence. Pseudomonas fluorescens was utilized as the positive control and Staphylococcus aureus as the negative control.
Lipase detectionNutrient agar supplemented with egg lectin was utilized to detect lipases. This medium is sensitive and is utilized specifically to detect lipase activity in bacteria. Media were inoculated by stabbing the agar and incubating it for 48h at 30°C. A positive result involves the formation of murky halos around the lipase-positive colony31. Bacillus subtilis was utilized as the positive control and Escherichia coli DH5-α as the negative control (Chávez D, personal communication).
Protease detectionProtease synthesis was evaluated in nutrient agar (Bioxon Cat. 210400, MD, USA) supplemented with 10% skim milk (Nido Kinder, Nestle, Mexico). The medium was inoculated by stabbing the agar and incubating it for 48h at 30°C. A halo around a colony indicates protease production31. B. subtilis was utilized as the positive control and E. coli DH5-α as the negative control (Chávez D, personal communication).
Amylase detectionAmylase synthesis was detected on nutrient agar with 1% starch. The medium was inoculated by stabbing the agar and incubating it for 72h at 30°C. To observe the results, 2ml of Lugol's iodine was added to the plates for 1min. Positive colonies exhibited clear halos31.
Esterase detectionEsterase activity was detected in nutrient agar supplemented with cholesteryl oleate and calcium chloride. The medium was inoculated by stabbing the agar and incubating for 72h at 30°C followed by 48h incubation at 4°C. Esterase positive bacteria exhibited accumulation of calcium at the edges of the colonies17.
Cellulose detectionCellulose-producing bacteria were detected on carboxymethylcellulose (CMC) plates. In this method, CMC plates were flooded with Gram's iodine. Gram's iodine forms a bluish-black complex with cellulose, but not with hydrolyzed cellulose, providing a sharp and distinct zone around cellulose-producing microbial colonies within 3–5min22.
Antifungal activity evaluationAntagonism experiments were performed against three phytopathogenic fungi belonging to the genera Aspergillus, Collechotrichum, and Fusarium, isolated from the Ataulfo variety of mango fruits from crops from the Miranda Fonseca community (17°13′25.1″N, 100°25′50.7″W) in Atoyac de Alvarez, Guerrero Mexico. A 3-mm cube of mycelial growth was placed on the center of a plate of potato dextrose agar (PDA). Next, four different bacterial isolates were inoculated 2.5cm from the center of the plates. The inoculated plates were incubated at room temperature for seven days. The plates were observed daily at 24h intervals, and the presence or absence of fungal growth inhibition was recorded31. Experiments were conducted in a completely randomized design with three replicates and four repetitions.
Molecular identification of PGPBBacterial DNA was extracted using the REDExtract-N Amp Tissue PCR Kit according to the manufacturer's instructions (Sigma-Aldrich, Cat. XNAT-100RXN, MO, USA). Then, using PCR, the 16S rRNA gene was amplified. The master mix was prepared with the following reagents 2.5μl PCR buffer (10×), 1μl MgCl2 (50mM), 1μl dNTP (10mM), 1μl of each primer at 10pmol/μl, 1μl Taq DNA polymerase (Thermo scientific)
3U/reaction, 17μl H2O mili-Q water, 2μl DNA (100ng) for a total volume of 25μl. The amplification protocol utilized was as follows: 1 cycle at 94°C for 3min, 30 cycles at 94°C for 1min, 54°C for 1min and 72°C for 2min, and a final extension at 72°C for 10min47. PCR products were mixed with SYBR® Green and loaded on a 1.5% agarose gel for electrophoresis (Molecular Probes, Invitrogen; Carlsbad, CA, USA). Positive results were indicated by a band of approximately 1500bp. The PCR products were washed, quantified, normalized, and sequenced directly with the Applied Biosystems BigDye Terminator v3.1 Cycle Sequencing kit and with the same primers utilized in the PCR reaction at the University of Arizona Genetics Core. Bidirectional sequencing was performed on an Applied Biosystems 3730xl DNA Analyzer (Foster City, CA, USA)16. The sequences were verified by inspecting the chromatograms in Sequencher (v. 4.5) (Gene Codes Corporation, Ann Arbor, MI). The sequence consensus was performed using ChromaSeq29 implemented on Mesquite v. 2.630. Phylogenetic analysis was performed on MEGA (v. 7.0)25. The sequences were grouped into operational taxonomic units (OTU) based on the sequence identity percentage and deposited in GenBank.
All the indigenous strains selected as PGPB were preserved in triplicate in 40% glycerol in an ultra-low temperature freezer for later studies.
ResultsSelection and isolation of bacterial morphotypesFrom a total of 125 samples, 151 morphotypes were obtained through selection according to their morphological differences. The samples obtained from the soil and rhizosphere isolates had the highest percentage of bacteria of interest with each category (31% of the isolates), while the leachate contained the lowest percentage (8% of the isolates; Table 1). We obtained 151 isolates of indigenous bacteria, which were preserved in glycerol in an ultrafreezer to use as a bank for future studies in agricultural crops in Guerrero, Mexico.
Screening of plant growth promoting-bacteria from mine tailings at El Fraile, Taxco, Mexico.
| Sample | Strain isolated (%) | Detection of mechanism of plant growth promotion (%) | ||||||
|---|---|---|---|---|---|---|---|---|
| Direct/biofertilization | Indirect/biocontrol | |||||||
| Fixation N2 | Phosphate solubilization | IAA | Gibberellins | Siderophores | Antifungal activity | Lytic enzymes | ||
| Soil | 31 | 17 | 5 | 2 | 17.8 | 17.8 | 16 | 25.1 |
| Rhizosphere | 31 | 19 | 3 | 10 | 19.2 | 25.2 | 7 | 27.8 |
| Root inner tissue | 17 | 12 | 0 | 0 | 6.6 | 3 | 2 | 7.4 |
| Leachate | 8 | 1 | 1 | 0 | 0 | 0 | 1 | 2 |
| Water | 13 | 3 | 2 | 0 | 5.4 | 2 | 1 | 4.6 |
| Total | 100% (151) | 52% (79/151) | 11% (17/151) | 12% (18/151) | 49% (74/151) | 48% (72/151) | 27% (41/151) | 66.9% (101/151) |
Only 52% (79/151) of the isolates could fix nitrogen after 24h. The most effective N2 fixators were the isolates obtained from rhizosphere and soil, with 19% (29/151) and 17% (26/151), respectively (Table 1 and Fig. 1A), and they corresponded to the genera Chryseobacterium, Bacillus, and Staphylococcus.
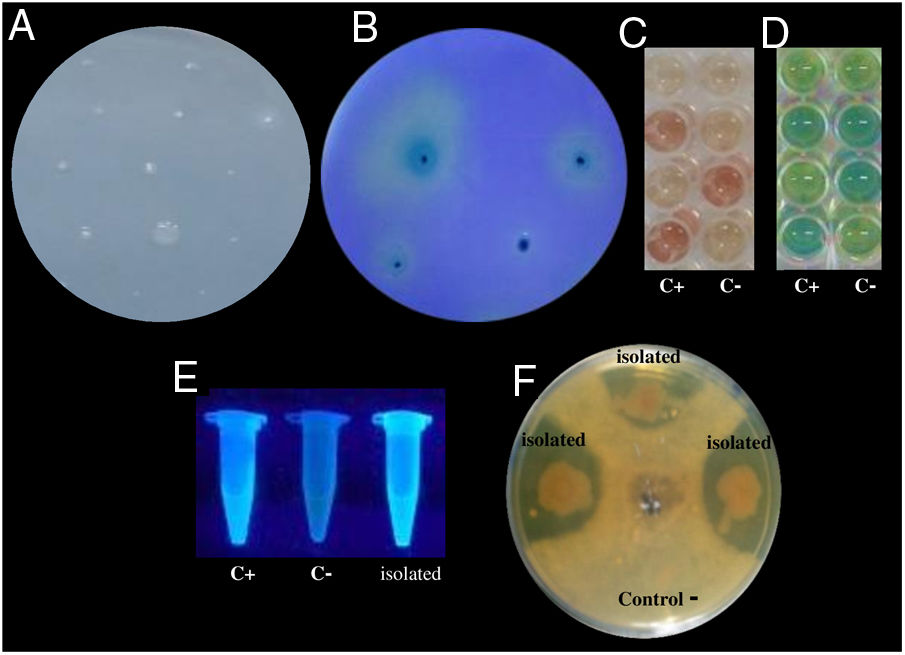
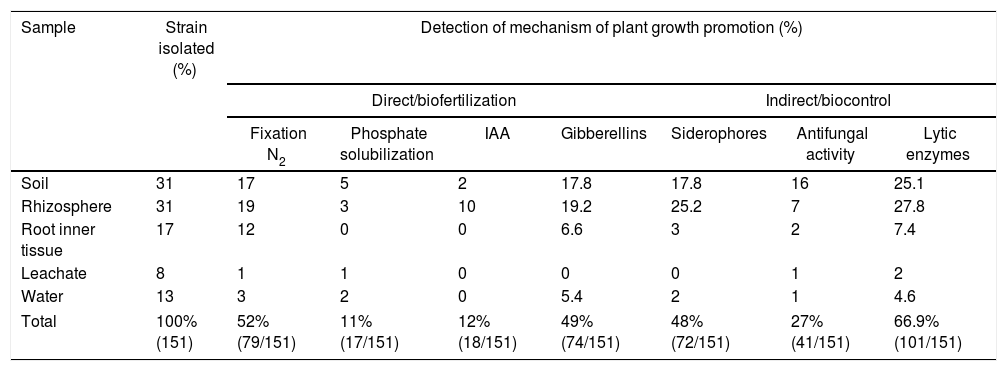
Examples of in vitro detection of plant growth promoting-bacteria and antagonistic characteristics. (A) Nitrogen fixation of different isolates; bacterial growth is indicative of nitrogen fixation. (B) Phosphate solubilization of several isolates; a clear halo around the colonies indicates the precipitation of phosphate. (C) Indoleacetic acid (IAA) production; the bottom wells contain the positive control (C+) A. vinelandii, and the negative control (C−) Salmonella Enteritidis, and the other wells contain the isolates. (D) Gibberellin detection; the bottom wells contain the C+, A. vinelandii, and C−, S. Enteritidis, and the other wells contain the isolates. (E) Siderophore detection; the C+ P. fluorescens is in the leftmost microtube, the C− S. aureus in the center, and the isolates on the right. (F) Antagonistic activity of different isolates (top, right, and left) against Fusarium sp., and the negative control (bottom).
In the evaluation of phosphate solubilization, only 11% (17/151) of the isolates were able to solubilize phosphate. These isolates were most frequently found in soil, with 5% occurrence rate (8/151) (Table 1 and Fig. 1B), and they corresponded to the genera Chryseobacterium, Bacillus, Staphylococcus, Serratia, Stenotrophomonas, and Acinetobacter. Phosphate solubilization is expressed quantitatively through the solubilization index (SI). The isolates obtained from water samples exhibited the highest SI, with a range of 7–10.1mm. The genera isolated from water included Stenotrophomonas and Acinetobacter. The mechanism of solubilization was determined by adding phosphatase activator (MgSO4); 77.7% (13/17) solubilized phosphate through enzymatic activity, 22.3% (4/17) by producing organic acids, and 22.3% (4/17) by utilizing both of these mechanisms.
IAA productionIAA production was exhibited by 12% (18/151) of the isolates from the rhizosphere, while the isolates from the inner root tissue, leachate, and water were unable to produce IAA. Isolates that exhibited the highest concentrations of IAA produced 16.5–43.2μg/ml (Table 1 and Fig. 1C). The IAA-producing isolates were identified within the genera Staphylococcus, Bacillus, Chryseobacterium, Pseudomonas, Curtobacterium, and Mycobacterium.
Gibberellin productionForty-nine percent (74/151) of the isolates were found to produce gibberellins. Samples from the rhizosphere contained 19.2% (29/151) of these samples, followed by soil with 17.8% (27/151) of the isolates (Table 1 and Fig. 1D). No isolates from the leachate samples produced gibberellins. The genera that synthesized this phytohormone included Staphylococcus and Bacillus.
Siderophore productionForty-eight percent (72/151) of the isolates produced siderophores. Most of these strains were isolated from the rhizosphere, with a total of 25.2% (38/151) of the strains, followed by isolates from the soil with 17.8% (26/151) (Table 1 and Fig. 1E). Isolates from the leachate could not produce siderophores. The genera that produced these metabolites corresponded to Chryseobacterium, Bacillus, Serratia, Curtobacterium, and Staphylococcus.
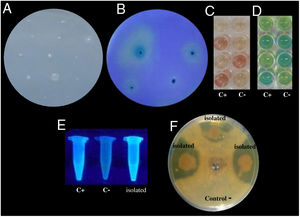
Antifungal activityAntifungal activity was evaluated against three phytopathogenic fungi from the genera Aspergillus, Collechotrichum, and Fusarium known to cause infections in Ataulfo mango from the coast of Guerrero, Mexico. Only 27% (41/151) of the isolates exhibited antifungal activity against at least one of the fungi. The greatest percentage of isolates that possessed antifungal activity were isolated from the soil with 16% (24/151), while only 1% of the isolates from water and leachate (2/151) were antagonistic toward the fungi (Table 1 and Fig. 1F). These isolates with antifungal activity were from the genera Pseudomonas, Serratia, and Bacillus. The isolate belonging to the genus Pseudomonas showed the strongest antifungal activity against Fusarium with 16.5% of strains showing antagonism (24/151) and the least against Colletotrichum with 10.0% (15/151).
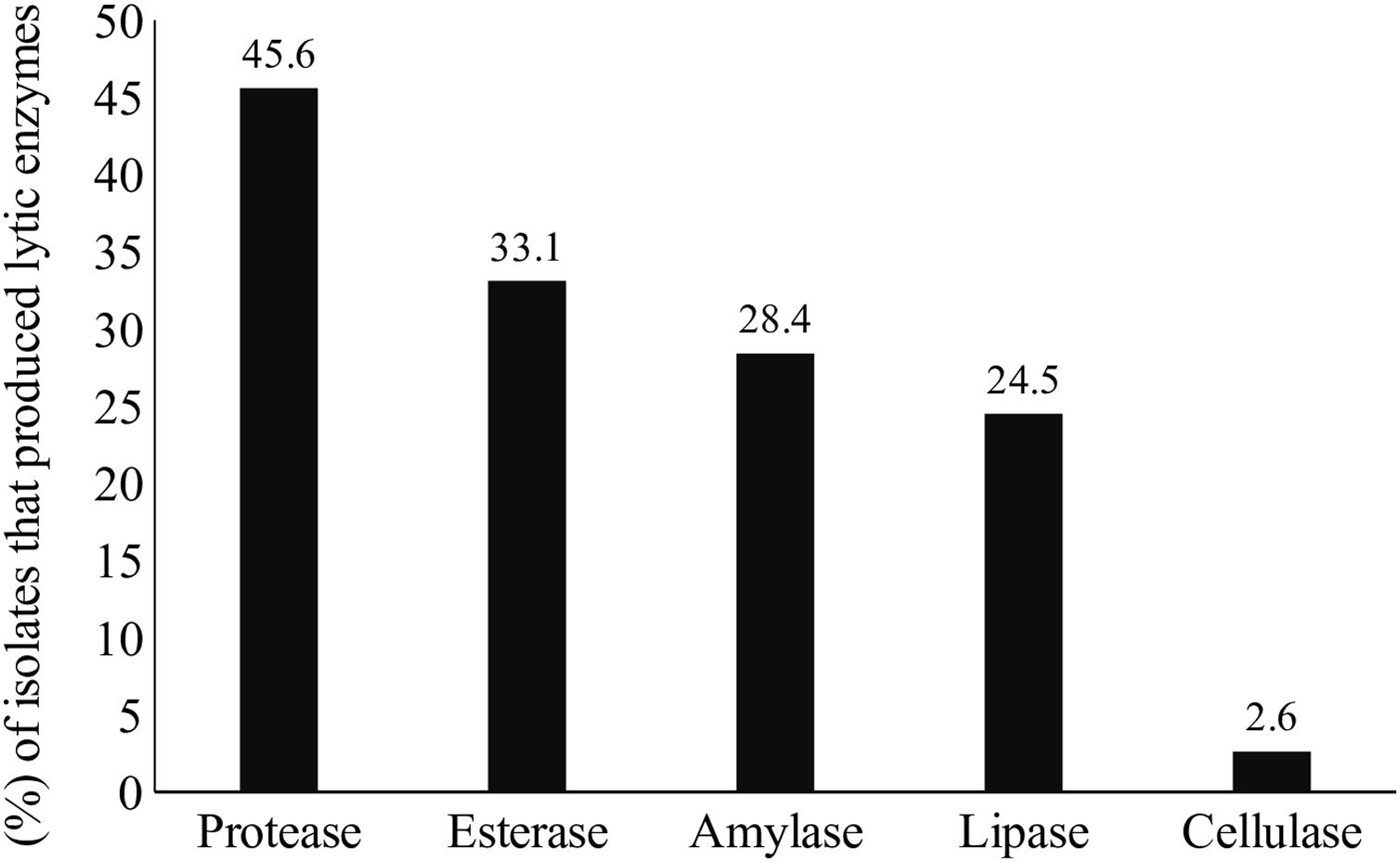
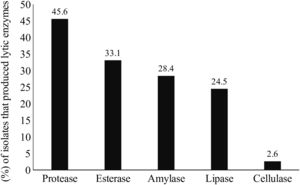
Lytic enzyme productionThe production of at least one class of lytic enzymes (including proteases, lipases, amylases, esterases, and cellulases) was observed in 66.9% (101/151) of the isolates (Table 1 and Fig. 2). The primary type of lytic enzyme produced was protease, followed by lipase, and the least common lytic enzyme produced was cellulase (Fig. 3).
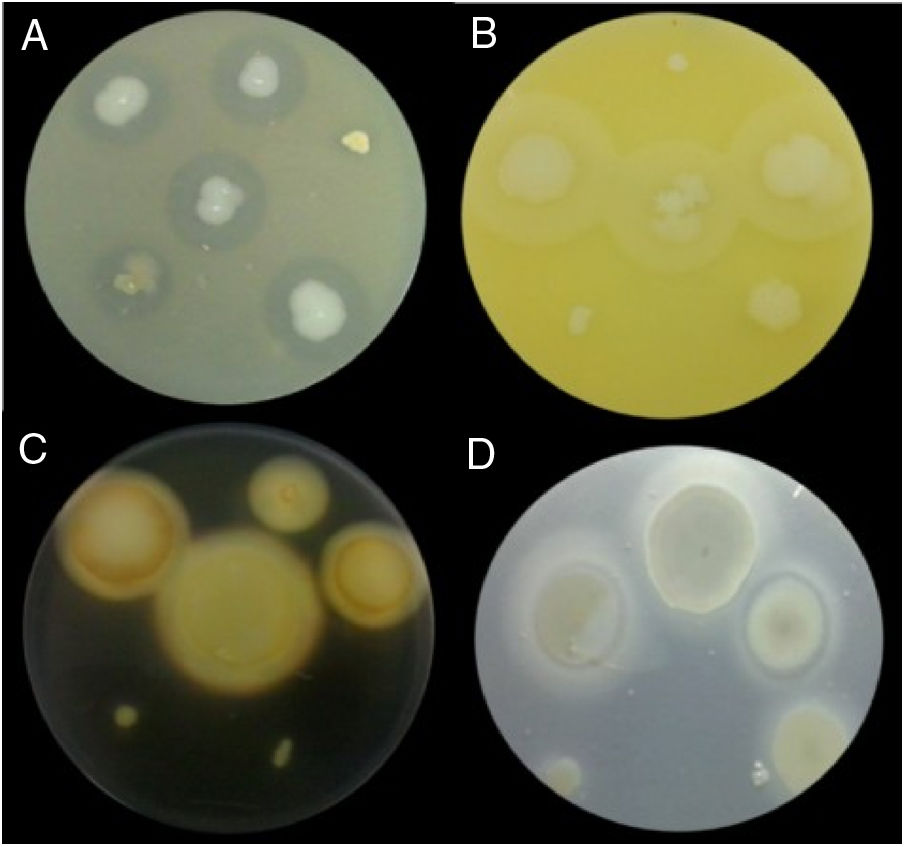
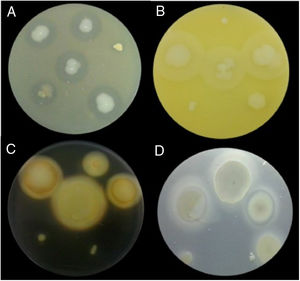
Examples of lytic enzyme production. (A) In skim milk medium, protease synthesis was indicated by a clear halo around the bacterial colony. (B) In egg lectin medium, lipase activity was indicated by a cloudy halo around the bacterial colony. (C) In the starch medium, amylase activity was indicated by a yellow halo around the bacterial colony. (D) In cholesteryl oleate and calcium chloride medium, esterase synthesis was indicated by a gray halo around the bacterial colony.
These kinds of metabolites have been shown to be involved in antifungal activity; therefore, we tested for a positive correlation between the production of each of the lytic enzymes and antifungal activity. Only lipases and esterases correlated with antifungal activity against species of Aspergillus.
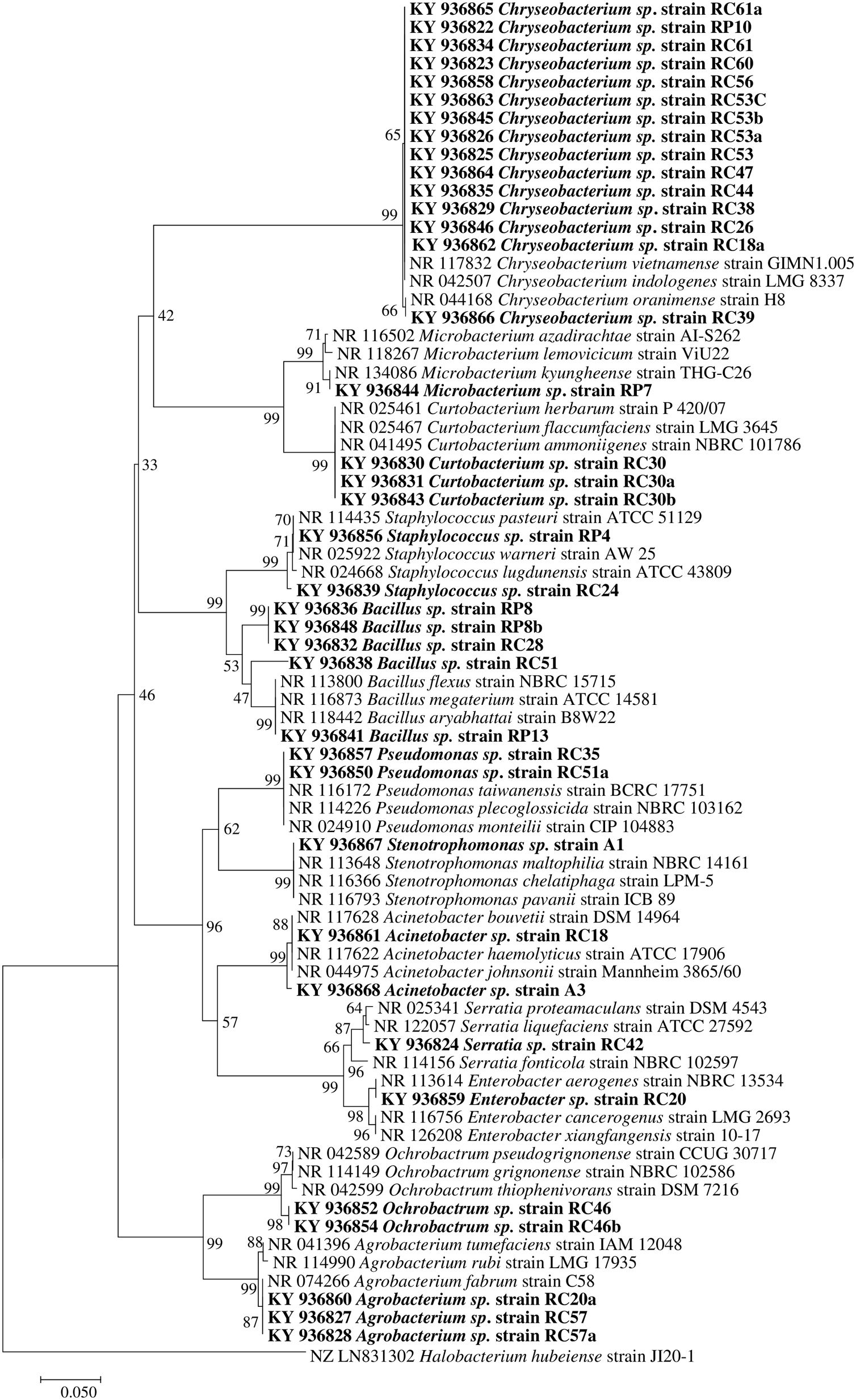
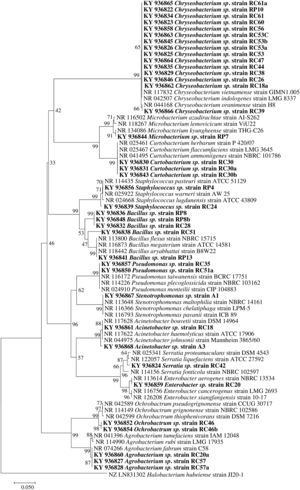
Molecular identification of plant growth-promoting bacteriaTwelve genera were identified through the 16S rRNA sequence analysis (Fig. 4) including species of Chryseobacterium, Bacillus, Pseudomonas, Mycobacterium, Staphylococcus, Curtobacterium, Serratia, Stenotrophomonas, Acinetobacter, Enterobacter, Ochrobactrum, and Agrobacterium.
Phylogenetic tree based on 16S rRNA nucleotide sequences of bacterial isolates from mine tailings. The evolutionary history was inferred using the neighbor-joining method38 with 1000 bootstrap replications test9 and the evolutionary distances were computed using the Jukes–Cantor method21. The sequences analyses were conducted with MEGA 7 software25.
Currently, there are few studies of the microbes isolated from mine tailings at El Fraile, Taxco, and their biotechnological potential is little known. Toribio-Jiménez et al.46 reported different bacterial isolates with capacity to produce biosurfactants and tolerate heavy metals. In our study, some bacteria isolated from different environmental matrices at the El Fraile tailing site are capable of promoting plant growth. It has been demonstrated that the presence of heavy metals in the soil decreases microbial biodiversity; however, the interaction among different organisms may play a key role in the survival of the species living under abiotic stress28.
The presence of bacteria in the rhizosphere may be attributed to chemotaxis due to the exudates secreted by roots that can modify the physiochemical properties of the soil, forming mutualistic associations in the rhizosphere2. Alternatively, plants that grow on the mine tailings are associated with rhizosphere microbial communities that play a key role in the modification of the availability and toxicity of the heavy metals for plants28. In response to iron limitation, these bacteria can secrete siderophores—low-molecular-weight ferric-specific ligands—allowing them to survive in soil contaminated with heavy metals26. A number of studies report that bacteria isolated from sites contaminated with heavy metals exhibit mechanisms that promote plant growth8. The biological fixation of nitrogen is the main process by which this macro-element is obtained, and the presence of atmospheric nitrogen fixators in the rhizosphere is due to the protective microenvironment and stimulants present in the roots that are found where the demand of nitrogen is greatest in plants33. De-Bashan et al.7 reported Bacillus pumilus capable of fixing nitrogen isolated from soil contaminated with heavy metals. Sarathambal and Llamurugu39 reported the genus Bacillus and Serratia as diazotrophs isolated from Bermuda grass (Cynodon dactylon). Singh et al.40 showed that Chryseobacterium increases nitrogen fixation by 50% in crops of Macrotyloma uniflorum. To the best of our knowledge, no previous research indicates that Staphylococcus can act as a diazotroph. With regard to the Agrobacterium genus, there are a few works that report diazotroph behavior5.
Only 11% (17/151) of the isolates could solubilize tricalcium phosphate, and they are distributed among all the environmental matrices except internal root tissue. The bacteria that are inside the root tissue (endophytes) do not solubilize this compound, since they are not in direct contact with the inorganic phosphate. Only the rhizobacteria are in contact with the inorganic phosphate, and they do solubilize it. In the leachate, the phosphate may remain undissolved due to its physiochemical conditions. Romero et al.37 determined that the leachate at El Fraile is pH 2.7, which could be facilitating the solubilization of phosphate ions. One mechanism for the dissolution of mineral phosphate by bacteria is the production of organic acids, which can chelate the cation bound to phosphate with their hydroxyl and carboxyl groups to be converted to soluble forms, and then become available to the plant4. Another mechanism employed by bacteria to solubilize phosphate is through the activity of acid phosphatase, a class of enzymes that catalyze the hydrolysis of phosphomonoesters at acidic pH4.
It was observed that the SI ranges from 7 to 10.1mm and corresponds to the genera Stenotrophomonas and Acinetobacter isolated from water. The SI values for A. vinelandii are greater than the value of 2.0 reported by Nautiyal34 in Bacillus polymyxa and 7.0 in P. fluorecens isolated from neutral soil. Islam et al.20 reported SI values ranging from 4.6 to 6.7mm in Acinetobacter and Klebsiella. In this study, Chryseobacterium was identified within the group of solubilizing bacteria. Singh et al.40 reported that this genus increases the solubilization of phosphate by 50%. All of these studies suggest that the type of microorganisms that solubilize phosphates varies from one environment to another.
Phytohormone synthesis by bacteria can strongly improve plant growth and development, as well as the plant response to the environment. Gibberellins are involved in seed germination, sprout emergence, floral induction, and fruit development. The production of gibberellins was more frequent in isolated rhizosphere bacteria, which could be due to the enriched microenvironment within the roots6. Pandya and Desai35 reported that an isolate of Pseudomonas monteilii capable of producing GA3 was isolated from the rhizosphere of Orotava sativa. The genera Chryseobacterium, Serratia, and Staphylococcus have not been previously reported to produce gibberellins.
IAA is the most abundant type of auxin. It can stimulate cellular elongation in bark, vascular tissue differentiation, growth, apical dominance, and lateral root initiation26. Here, we found that IAA-producing isolates were from the genera Bacillus, Pseudomonas, Chryseobacterium, Curtobacterium, Staphylococcus, and Mycobacterium. Ten percent of these isolates are in the rhizosphere and produce 6.2–43.2μg/ml, compared to the production of 5.7μg/ml by the positive control A. vinelandii. Similarly, Yu et al.49 reported 13 bacterial genera isolated from soil in the V–Ti magnetite mine. Among these species, B. pumilus and B. subtilis were isolated and observed to produce IAA at concentrations ranging from 2.2 to 83.05μg/ml. Eleven of the species produce more than 60μg/ml. In contrast, Islam et al.19 isolated the genera Lysinibacillus, Bacillus, and Pseudomonas from neutral soil that could produce 3.1, 2.1, and 2.3μg/ml of IAA, respectively. It is critical to understand that the production of IAA in bacteria varies among species and strains. This production is influenced by their growth conditions, growth phase, and substrate availability. There are data suggesting that many types of bacteria produce IAA. It has been observed that more than 80% of the bacteria isolated from the rhizosphere can synthesize IAA23.
Iron-limited conditions favor the synthesis of siderophores, and this characteristic serves a key role in the antagonism of phytopathogenic biocontrol by limiting iron availability1. In this study, we found that 48.3% of the isolates could produce siderophores. Talavera et al.42 reported elevated concentrations of Fe2+ in the mine tailings at El Fraile, ranging from 2.49 to 25mg/g. This finding suggests that siderophore production by bacteria isolated could be linked to the exposure to high levels of iron in the environment. It has been demonstrated that siderophore-producing bacteria improve the chlorophyll content and growth of a variety of plants thriving in soils contaminated with heavy metals48. The isolation of Serratia was consistent with the results reported by Koo and Cho24, who isolated siderophore and IAA-producing bacteria of the genus Serratia strain SY5, from Echinochloa cruzgalli roots exposed to soil contaminated with metals and petroleum. In addition, when SY5 was inoculated in vitro into the roots of Zea mays in soil contaminated with Cd, a significant increase in root biomass was observed. Yu et al.49 reported B. subtilis, B. pumilus, Rhizobium, and Ochrobactrum intermedium to be the most frequently isolated bacteria from soil in the V–Ti magnetite mine. Kumar et al.26 isolated Bacillus thuringiensis, Azotobacter chrococcum, P. imensis, and Pseudomonas pseudoalcaligenes, which produced siderophores, IAA, hydrogen cyanamide and dissolved phosphate from soil contaminated with industrial residues and heavy metals. To the best of our knowledge, no previous studies indicate that the genera Agrobacterium, Chryseobacterium, and Staphylococcus can produce siderophores. This report is the first to describe these characteristics for isolates obtained from contaminated sites.
The genera found to possess antifungal activity included Pseudomonas, Serratia, and Bacillus, which suggests that antifungal activity is mediated by secondary metabolite production more than by the synthesis of lytic enzymes and siderophores. George et al.10 reported that an isolate of Pseudomonas aeruginosa from the soil of a tea plantation exhibited antifungal activity against ten phytopathogens, including Aspergillus, Fusarium, and Colletotrichum. It was demonstrated that this antifungal activity is a consequence of the production of metabolites including 1-hydroxyphenazine, pheneizine, and pyocyanine. Idris et al.18 reported that antifungal activity against Fusarium is due not only to antifungal compounds but also to other metabolites, such as siderophores, hydrogen ions, ethylene, hydrocyanic acid, and ammonia. Bacillus and Pseudomonas have been reported to be antagonists of different phytopathogenic fungi relevant to agriculture41.
ConclusionsSeveral PGPB that also had phytopathogenic characteristics were isolated from soil and plant materials from El Fraile mine tailings. The beneficial characteristics of these PGPB included phytopathogenic fungal antagonists, production of metabolites of interest such as phytohormones and lytic enzymes, as well as nitrogen fixation and mineral solubilization. These bacteria are potentially very useful for applications in the areas of agronomy, biotechnology and as a booster of phytoremediation of contaminated soils and should be further explored in the near future.
Conflicts of interestThe authors declare no conflict of interest.