The Riachuelo river basin (RRB) is considered one of the most polluted environments in the world. Knowledge of arbuscular mycorrhizal fungi (AMF) adapted to this extremely polluted environment is important for the establishment of future soil restoration projects. This work aims to make a first list of AMF species present on the RRB. Soil and root samples were randomly taken in an area of approximately 1500m2, mycorrhization percentages were evaluated. AMF species were detected by molecular and morphological techniques. Sixteen AMF morphological species and 64 molecular species were reported in this work. Dominikia iranica, Funneliformis constrictum, Funneliformis mosseae, Rhizophagus intraradices, Rhizophagus irregularis and Septoglomus viscosum were detected by both techniques while Claroideoglomus sp. was only detected by pyrosequencing. The list of species reported in this work represents the first description of the RRB AMF community.
La cuenca del río Riachuelo (CRR) es considerada uno de los ambientes más contaminados del mundo. Conocer los hongos formadores de micorrizas arbusculares (HFMA) adaptados a este ambiente extremadamente contaminado es importante para el establecimiento de futuros proyectos de restauración de suelos. Este trabajo se propuso hacer una primera lista de especies de HFMA presentes en la CRR. Se tomaron muestras de suelo y raíces al azar en un área de aproximadamente 1500m2 y se evaluaron los porcentajes de micorrización. La identificación de especies de HFMA se basó en técnicas moleculares y morfológicas. Se detectaron 16 especies morfológicas y 64 especies moleculares de HFMA. Dominikia iranica, Funneliformis constrictum, Funneliformis mosseae, Rhizophagus intraradices, Rhizophagus irregularis y Septoglomus viscosum se detectaron mediante ambas técnicas, mientras que Claroideoglomus sp. solo fue detectado por pirosecuenciación. La lista de especies reportada en este trabajo representa la primera descripción de la comunidad de HFMA de la CRR.
One of the most important concerns raised by the world society is the progressive chemical degradation of soils by contamination as a consequence of human activities. The Matanza and the Riachuelo are lowland rivers that run through the Pampean plain along the South metropolitan area of Buenos Aires (Argentina). The Matanza-Riachuelo basin is the most important water system in this area (233800 Ha), covering 14 municipalities of Buenos Aires province and the city of Buenos Aires. According to the last census (2010), about 8 million people live in this area (https://www.indec.gov.ar, last accessed: February 2019). The lower portion of the Riachuelo river basin (RRB) is the most physically, chemically and biologically degraded environment of the Matanza-Riachuelo river basin. The high population and industrial density were the causes of the extreme deterioration of the basin water, air and soil. Nowadays this area is considered one of the ten most polluted environments in the world2. A wide range of inorganic and organic pollutants can be found in the area. Among them, the most commonly found are: As, Cd, Cr, Cu, Hg, Fe, Ni, Pb, Zn and Mn12.
Successive attempts have historically been made to restore the environment of the RRB, with particular emphasis placed on the aquatic environment ecosystem (http://www.acumar.gob.ar/eje-ambiental/, last accessed: February 2019)). For the edaphic environment, tasks of landscape restoration were carried out: the towpath space was recovered (previously occupied by industries and precarious housing), visible wastes were removed, and native plants were planted. No attempt to remedy the basin soil has been made yet.
Among the available technologies to remediate polluted soils, bioremediation involves lower costs and minimal environmental alterations. Arbuscular mycorrhizal fungi (AMF), belonging to Glomeromycota Phylum, were previously found in soils with high contents of heavy metals (HM), proving that they have established mechanisms of resistance or tolerance to these adverse conditions9. AMF are strict biotrophs, needing to colonize a host root to get access to carbohydrates and complete their life cycle. In exchange, fungi improve host tolerance to biotic or abiotic stressful conditions among other benefits. The vast majority of plants are capable of establishing a symbiosis with AMF, increasing the volume of explored soil by roots through the establishment of hyphal networks. They also contribute to soil aggregation improving the edaphic structure, being considered key microorganisms for the productivity and diversity of ecosystems14. Based on this fact, the symbiosis establishment could be considered a plant strategy to improve its status in highly polluted environments. Recently it was confirmed that AMF colonization improves the efficiency of rhizoremediation because of the uptake of contaminants by fungal hypha and their transport to the plant tissues9.
Knowing the AMF species adapted to the extremely polluted environment of the RRB is important for future soil restoration projects. Species characterization by the traditional approach (based on propagule traits) has many challenges: the degraded conditions of spores in the field, the presence of non-sporulating species or the difficulty of distinguishing between functional and morphological diversities, among others. Second-generation sequencing technologies have been increasingly considered to be useful and complementary tools to elucidate the diversity of AMF in environmental samples4.
The aim of this work was to make a preliminary list of the AMF species presenting the RRB by molecular and morphological techniques, and to evaluate the degree of colonization of the plant roots growing there.
Sampling took place in the lower portion of the RRB (Avellaneda, 34°39′27.36″S, 58°22′34.37″W) in September 2013. Twenty-five soil cores (10cm depth) were randomly taken in an area of 1500m2 and then homogenized into a unique sample in order to remove the effects of field heterogeneity1. The soil sample was divided and stored at 4°C for subsequent physicochemical analyses, evaluation of HM content and identification of AMF species by molecular and morphological techniques.
A survey of the local flora was carried out and plants were collected from the towpath (0.5–15m from the water edge). Seventeen plant species were identified: Festuca arundinacea, Paspalum distichum, Sorghum halepense, Taraxacum officinale, Senecio bonariensis, Chamaemelum nobile, Trifolium sp., Cyperus eragrostis, Dichondra microcalyx, Salix babylonica, Morusnigra, Ricinus communis, Commelina erecta, Hydrocotyle bonariensis, Conium maculatum and Juncus pallescens.
Roots of herbaceous mycorrhizal plants were sampled to evaluate their mycorrhizal status. They were cleared in KOH (10%) and stained with trypan blue in lactic acid (0.02%)11. Thirty root segments (1cm) per plant were examined under a light binocular microscope (model: Olympus BX51) in order to estimate the percentage of AMF colonization6. Mycorrhization frequency was calculated as the percentage of root segments containing AMF hyphae, arbuscules, coils or vesicles; mycorrhization intensity was estimated by sorting out the root segments in percentual intervals of AMF intraradical cover. The percentage of arbuscules, coils and vesicles, were separately registered, considering arbuscules and coils as characteristic structures of an active symbiosis and vesicles as storage structures3. Mycorrhization frequency reached high values (72.2%, SD: 31.7); however, the intensity of AMF colonization did not (38.2%, SD: 19). Arbuscules and coils were more commonly found than vesicles, being detected in 86.5% (SD: 26.2) and 31.1% (SD: 23) of the mycorrhizal roots, respectively. Intraradical AMF spores were observed, though in a very low proportion. Typical mycelium and structures of Glomus tenue were observed associated with J. pallescens roots. AMF colonization was not only frequent but also in an active stage due to the high percentage of arbuscules and coils. Mendoza et al.8 have also reported the presence of one of these mycorrhizal plants (H. bonariensis), not only in the lower portion of the basin but also in the middle and upper portions. The authors observed that H. bonariensis plants were always colonized by AMF.
Physicochemical analyses of the soil and the evaluation of HM contents were conducted at the Instituto de Geocronología y GeologíaIsotópica (INGEIS, CONICET) and at the Comisión Nacional de Energía Atómica (CNEA, Regional Cuyo unit) (Table 1; supplementary material). HM can be naturally found in the soil or originated as a result of anthropogenic activities. Many of these heavy metals are essential micronutrients for plant growth; however, when exceeding certain concentrations, these elements may become toxic and hinder plant growth. High contents of Fe, Zn, Pb, Cu, Cd, Sr, Ni, Cr and Mn were found in the RRB soil (Table 1; supplementary material). However, according to the Argentine legislation (Law 24051: Hazardous waste in Argentina, decree 831/93, Annex II, Table 9), only Cd, Cr and Mo exceed the levels considered suitable for agricultural and residential use of the soil. Other authors have previously reported similar levels of soil contamination across the Matanza-Riachuelo river basin by many of the HM reported in this paper8,12.
Twelve pot trap cultures were established to facilitate the taxonomical identification of AMF spores. One liter pots were were filled with a sterile (100°C for 1h, three consecutive days) perlite:soil mixture (2:1) as growing substrate and 10g of the RRB sampled soil containing AMF propagules and mycorrhized root segments as inoculum. Sorghum, tomatoes, peas and clover were used as host plants. Cultures were monitored every two months for eighteen months. AMF propagules were isolated from 100g (dry weight) of RRB soil and from trap culture substrate by wet sieving and decanting method. Spores that appeared healthy were manually selected and removed using a micropipette under a stereomicroscope (model: Olympus SZ61). All propagules were mounted on slides containing polyvinyl alcohol-lactic acid-glycerol (PVLG) and a mixture of PVLG-Melzer reagent. Spore and subcellular structure characteristics were observed under a light binocular microscope (model: Olympus BX51). Taxonomic classifications, whenever possible, were made in accordance with the descriptions available at the International Culture Collection of Vesicular Arbuscular Mycorrhizal Fungi (INVAM) website (http://invam.wu.edu, last accessed: February 2019) and at Dr. Blaszkowski's website (http://www.zor.zut.edu.pl/Glomeromycota/, last accessed: February 2019). Taxonomic assignments were done according to the Index Fungorum (http://www.indexfungorum.org/names/names.asp, last accessed: February 2019).
Propagules of 15 different AMF species were isolated from RRB sampled soil and pot trap cultures and classified as: Acaulospora sp., Dominikiairanica, Entrophospora infrequens, Funneliformis constrictum, Funneliformis mosseae, Gigaspora sp., Rhizoglomus microaggregatum, Rhizophagus sp., Rhizophagus clarus, Rhizophagus diaphanus, Rhizophagus intraradices, Rhizophagus irregularis, Sclerocystis sinuosa, Scutellospora pellucida and Septoglomus viscosum.
DNA was isolated from 0.25g (dry weight) of RRB soil (previously sieved through a 2mm mesh) in triplicate. The MoBio Power Soil DNA isolation kit was used, following the manufacturer's instructions (Mo Bio Laboratories, Inc., Carlsbad, CA, USA). Amplification was performed on a Fast Start High Fidelity PCR system (Roche Applied SCIENCE, Mannheim, Germany) following the manufacturer's instructions. PCR conditions are detailed as follows: 95°C (5min); 30 cycles of 95°C (45s), 57°C (45s), 72°C (60s); 72°C (4min). AMV4.5F/AMDGR oligonucleotides were chosen because of their specificity to amplify AMF DNA in environmental samples4. Primers were specifically designed for 454 pyrosequencing with the Genome Sequencer-FLX (GS-FLX) 454 Titanium System (Roche Applied Science, Mannheim, Germany). Amplification products were purified using AMpure Beads (Agencourt) in order to remove primers, impurities and small spurious products. Amplification and 454 amplicon pyrosequencing were performed at the Instituto de Agrobiotecnología de Rosario (INDEAR), following Roche 454 software manuals.
Treatment of the obtained sequences was carried out according to Colombo et al.4 MOTUs were defined at 97% (97% dataset) and 90% (90% dataset) of sequence similarity. The first percentage was chosen in accordance to the traditional definition of microbial ‘species’. The second and lowest percentage was chosen due to the possible overestimation of AMF richness when considering a 97% similarity, as a high genetic diversity was observed within a single individual in comparison with other organisms, probably due to the asexual and multinucleated nature of AMF14. Representative sequences from each MOTU were compared with 18S rRNA published sequences on the MaarjAM (specific database for AM fungal) and the NCBI (National Centre for Biotechnology Information) databases using the nr/nt BLAST (Basic Local Alignment Search Tool) algorithm. Only sequences with query coverage and similarity values higher than 97% (E-values equal or close to zero) were considered. After MOTU identification, non-Glomeromycota sequences were removed from the data sets. Taxonomic assignments of Glomeromycota sequences were made according to the Index Fungorum. Whole analyzed sequences are available as FASTQ at the NCBI Sequence Read Archive database (accession number: SRR2968218).
A total of 179286 sequences (mean length: 204.8pb, SD: 16.1pb) were obtained and grouped into 217 MOTUs (97% dataset) or 92 MOTUs (90% dataset). According to the best BLAST hit in MaarjAM and the NCBI databases, 46.13% (64 MOTUs) and 48.41% (12 MOTUs) of the total reads belonged to Glomeromycota sequences, at the 97% and 90% datasets respectively (Table 2; supplementary material).
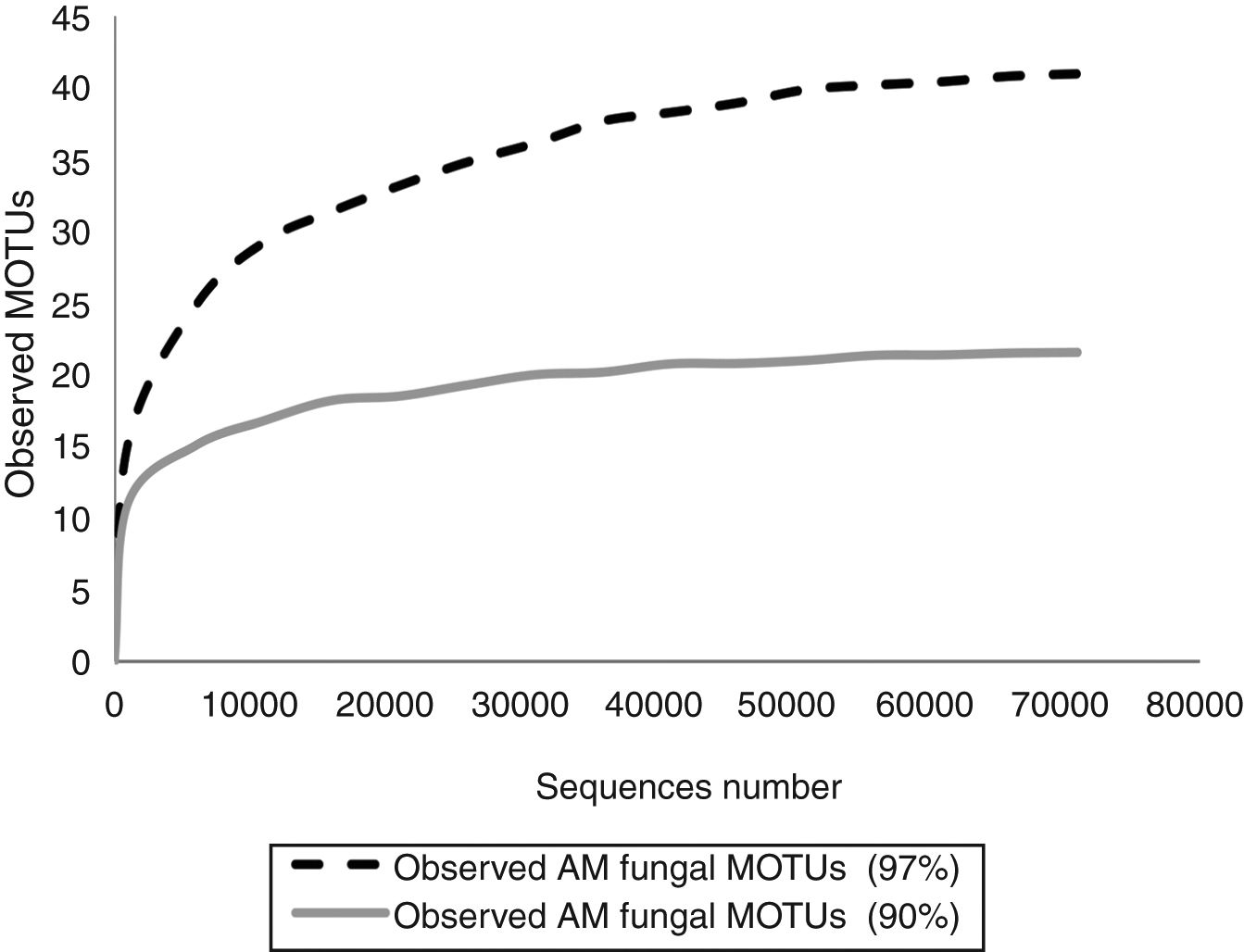
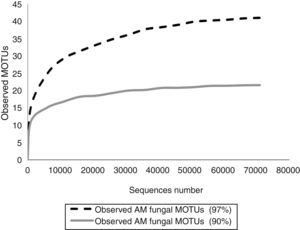
In order to know if the sequencing effort was enough to cover the entire AMF diversity, rarefaction curves were constructed with Quantitative Insights Into Microbial Ecology (QIIME) pipeline by randomly selecting a series of different sized subsets of reads from the data set. Each sampled subset was replicated ten times by the program. Rarefaction curves reached the asymptotes in both cases (Fig. 1).
Both datasets presented AMF MOTUs that could only be classified at the Phylum level. MOTUs belonging to the Order Glomerales were always the most abundant (95.9% and 91.1%), while Order Diversisporales sequences were only registered at 97% dataset (0.03%). AMF sequences of genera Claroideoglomus (36.1%), Dominikia (26.5%) and Rhizophagus (14.2%) were the most representative ones at 97% dataset whereas 15.8% of Glomerales sequences could not be classified at the genus level (Table 2; supplementary material). At 90% dataset the genus Claroideoglomus was also the most abundant (34.3%), followed by S. viscosum AMF species (3.1%). No sequence of the genus Rhizophagus was detected; however, 53.8% of the Glomerales sequences were not identified at the genus level in this dataset (Table 2; supplementary material). The higher abundance of Glomerales could be related to a higher tolerance of this order to extreme conditions due to their life history strategies, characterized by short life cycles, a great investment in spore production and the capability to interconnect different mycelia through hyphal bridges14.
In order to check the right taxonomic assignment of MOTUs, phylogenetic analyses were performed using the maximum likelihood distance method. Glomeromycota sequences from both molecular data sets were aligned using the ClustalW program. Bootstrap analysis with 1000 replicates was used to test the confidence of the branches. Phylogenetic trees were constructed and edited by the Tree Explorer of the MEGA 6.06 program. Both trees resulted in consistent ordinations of MOTUs. While the statistical supports of the branches were not always high, phylogenetic groups were well established, with the exception of the sequences belonging to F. constrictum and S. viscosum and the phylogenetic group formed by R. intraradices and R. irregularis sequences, which could not be distinguished in this analysis (data not shown).
Fifteen morphological species and 64 MOTUs of AMF were reported in this work. These results represent the first description of the AMF community of the extremely polluted RRB soil. Mendoza et al.8 detected AMF spores in the lower portion of the basin in Avellaneda city; however, they were not taxonomically identified. D. iranica, F. constrictum, F. mosseae, R. intraradices, R. irregularis and S. viscosum were the only species detected by both assessed techniques. The 90% dataset failed to identify MOTUs at the species level, except for F. mosseae and S. viscosum. Contrary to our expectations, AMF richness estimated by the 90% dataset was much lower than that detected by the morphological technique. Hence, the use of 90% similarity among sequences to delimitate MOTUs when studying AMF communities would not be advisable.
Claroideoglomus sp. MOTUs were abundantly detected by the pyrosequencing technique; however, spores were not recovered from the soil. This could be attributed to the non-sporulation of Claroideoglomus mycelium under the particular environmental conditions of RRB. Panwar et al.10 observed a variation in the sporulation dynamic associated to physico-chemical-climatic conditions of soil. However, Claroideoglomus spores were not recovered from the trap cultures either. In turn, E. infrequens and R. microaggregatum spores were recovered from RRB soil samples but their sequences were not. In a previous work, Colombo et al.4 reported the same inconsistency. The authors related the molecular undetectability of these species to the difficulty to isolate DNA from very small spores (R. microaggregatum) or to the low number of propagules in the soil (E. infrequens).
Many of the AMF species described in this work had been previously morphologically or molecularly detected in soils with high HM concentrations. Spores of F. mosseae were found in strongly contaminated soils with Pb, Zn and Cd in Shipham (England) and with Pb, Zn, Cu, Ni and Cd in Braunschweig (Germany), where Claroideoglomus claroideum was also abundantly found5. We detected F. mosseae in RRB soils by morphological and molecular techniques; however, spores of C. claroideum were not isolated in this sampled site. The genus Claroideoglomus was only detected by the metagenomic analysis of soil, but this technique failed to differentiate sequences at the species level. R. intraradices, R. clarus and C. claroidedum had been previously described in soil contaminated with Cd, together with Funneliformis geosporum, Funneliformis coronatum and Diversispora versiforme13; however, these three latter AMF species were not present in the sampled RRB site. Spores of R. intraradices were also found in soil containing high concentrations of Zn7. Typical Gigasporaceae auxiliary cells and G. tenue characteristic mycelium and vesicles were observed in association with root segments sampled in Ni contaminated soils15.
Knowing AMF species of the RRB would facilitate future isolation and conservation works of these fungal strains in order to use them in bioremediation assays.
Conflict of interestAuthors have no conflict of interest.
FundingThis work was supported by Universidad de Buenos Aires (UBA), Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET) and Agencia Nacional de Promoción Científica y Tecnológica (ANPyCT).
Authors thank to Dr. Pérgola Marina and Dr. Marta Cabello for their technical assistance, to Dirección de Flora y Fauna and Ministerio de Asuntos Agrarios (Buenos Aires province) for the extended sampling licenses and Autoridad de la Cuenca Matanza Riachuelo (ACUMAR) crew for their counselling.