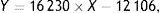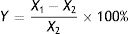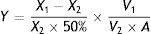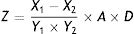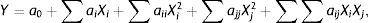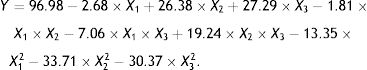The mycotoxin Ochratoxin A (OTA) causes serious health risks and is found in food products throughout the world. The most promising method to detoxify this compound is biodegradation. In this study, Aspergillus oryzae strain M30011 was isolated and characterized based on its considerable capacity to degrade OTA. The degradation product (compound I) of A. oryzae-treated OTA was isolated, and its toxicity response was also evaluated. Furthermore, the relationships between three key cultivation condition factors affecting the OTA degradation rate were examined using the response surface methodology (RSM). Compound I was identified as ochratoxin α (C11H9O5Cl), and the toxicity response experiments indicated that A. oryzae detoxified OTA to a great extent. A maximum degradation rate of 94% was observed after 72h. This study demonstrates the potential for using A. oryzae to detoxify OTA and suggests that it could be applied in the food industry to improve food safety and quality.
La ocratoxina A (OTA) es una micotoxina que pone en grave riesgo la salud; dicha toxina se encuentra en productos alimentarios a nivel mundial. El método más prometedor para reducir su toxicidad es la biodegradación. En este estudio se aisló la cepa M30011 de Aspergillus oryzae sobre la base de su considerable capacidad de degradar la OTA, y se efectuó su caracterización. Se aisló el principal producto de la degradación de la OTA (compuesto I) tratada con dicho microorganismo, también se evaluó la respuesta del hongo en términos de toxicidad. Además, se examinaron las relaciones entre tres factores claves de la condición de cultivo que afectan la tasa de degradación de la OTA mediante la metodología de superficie de respuesta (RSM). El compuesto I fue identificado como ocratoxina α (C11H9O5Cl) y los experimentos demostraron que A. oryzae M30011 redujo la concentración de OTA en gran medida. Se observó una tasa de degradación máxima del 94% transcurridas 72h. Este estudio demuestra el gran potencial de A. oryzae M30011 para reducir la toxicidad de la OTA y sugiere que podría aplicarse en la industria alimentaria para mejorar la seguridad y la calidad de los alimentos.
Ochratoxin A (OTA) is a fungal secondary metabolite that can contaminate agricultural products and poses severe global health risks27,38,39. Thus, considerable precautionary measures have been taken to control fungal OTA production and prevent the mycotoxins from contaminating foods and entering food chains. Despite these efforts, the detoxification of OTA-containing agricultural products is still a critical concern.
Several physical and chemical strategies are available for detoxifying OTA4,5,20,30,41,42. However, the efficacy of physical strategies is limited, and chemical strategies pose potential safety risks themselves. Most physicochemical methods are impractical for degrading OTA because either their application in the food industry is limited due to incomplete toxin degradation or they also lead to secondary pollution. In contrast, biological methods do not reduce the nutritional value of foods and exhibit high efficacy and specificity for detoxifying OTA1,6,13. Consequently, research efforts have recently focused primarily on biological detoxification methods.
Microorganisms have been specifically shown to detoxify OTA, and biological approaches such as these have broad application prospects28. However, some of the detoxification effects of various bacterial strains are actually the result of adsorption rather than degradation. Further, different degradation rates have been reported for different OTA-degrading strains11,15,23,24,29. Nevertheless, the potential for using OTA-degrading microorganisms in the food industry is lessened by several factors including long degradation times, incomplete degradation, and their sensitivity to fermentation products that are typically present in foods. Moreover, certain OTA-degrading strains may produce hazardous substances under varying conditions that can also increase health risks.
Most strains that exhibit strong OTA degradation ability are members of the bacterial genera Rhizobium, Bacillus, and Acinetobacter in addition to the fungal genera Aspergillus, Penicillium, and Rhizopus32. Despite the potential for microbial degradation of OTAs, strains that can efficiently degrade OTA are scarce, most of them exhibiting degradation times of one to two weeks2,26. Indeed, most of the reported OTA-degrading strains eliminate OTA toxicity by biosorption. Thus, strains that can efficiently detoxify OTA by biodegradation are rare37.
In addition to the above efforts, optimal conditions for OTA biodegradation have been evaluated including fermentation media components and individual or combined cultivation conditions, among others. These studies have contributed to the development of several successful microbial enzymatic degradation strategies3,17. The traditional “one-factor-at-a-time” method is generally used to optimize degradation conditions. However, this strategy is not only time consuming but also often leads to the incomplete examination of interacting factors, resulting in relatively weak predictions of optimal conditions. To overcome these limitations, the response surface methodology (RSM) and experimental factor design can be used to optimize process conditions while performing minimal numbers of experiments16.
Based on the above observations, the objectives of this study were to (i) screen and isolate OTA degrading strains from soils using safe deficient-media; (ii) examine the OTA degradation abilities of the recovered strains by thin-layer chromatography (TLC) and high-performance liquid chromatography (HPLC); (iii) identify and characterize the strain(s) by morphological identification, electron microscopy, and molecular analysis; (iv) analyze the OTA degradation products of the strains; and (v) optimize conditions for OTA biodegradation and evaluate the possibility of applying the strains in food and agricultural settings.
Materials and methodsChemicals and mediaOTA standard and M290 purification columns (for evaluating adsorption and the concentrations of toxins in the samples) were purchased from Pribolab (Pribolab Ltd., Singapore). All other chemicals were obtained from Banxia (Banxia Biological Technology Co., Ltd., China). The deficient medium used to culture organisms contained 0.25g/l KH2PO4, 1.0g/l NH4NO3, 0.25g/l MgSO4·7H2O, 0.001g/l FeSO4, 20% agar, and 1g/l l-phenylalanine or coumarin (pH 7.0). The basal medium contained 3g/l beef extract, 5g/l, soy peptone, 10g/l NaCl, and 6g/l glucose (pH 7.0). The isolated strains were cultivated in liquid medium (3g/l beef extract, 5g/l soy peptone, 10g/l NaCl, 6g/l, glucose, pH 7.0) to evaluate OTA degradation.
Strains exhibiting OTA degradation ability were stored in Potato Dextrose Agar (PDA) medium (comprising 200g/l potato, glucose 20g/l, 15g/l agar, pH 7.0). Identification of strains was conducted on Czapek–Dox Agar (CA) medium (3g/l NaNO3, 1g/l KH2PO4, 0.5g/l MgSO4·7H2O, 0.5g/l KCl, 0.01g/l FeSO4, 30g/l sucrose, 15g/l agar, pH 7.0), Czapek yeast extract (CYA) agar medium (1g/l K2HPO4, 10g/l Chamberlain concentrate, 5g/l yeast extract, 30g/l sucrose, agar 15g/l, pH 7.0), and malt extract agar (MEA) medium (20g/l malt extract, 1g/l peptone, 20g/l glucose, 15g/l agar, pH 7.0).
Strain isolationOTA-degrading strains were isolated from 100 samples including garden, swamp, and forest soils taken from different regions of China. Top soils were removed and 1kg of soil was taken from soil at a depth of 10–20cm by coning and quartering. One gram of each sample was added to 9ml of sterile water and mixed by vortexing for 30min. Samples were then diluted to approximately 1:1000 or 1:10000 in sterile water. Each dilution (1.0ml) was spread on the deficient solid medium and incubated at 30°C or 37°C for 24h. Colonies were subcultured three consecutive times under the same initial incubation conditions. To preserve the strains, they were cultured on Luria-Bertani (LB) media with sterile glycerol (20%) supplemented and stored at 20°C. Fungi were cultured on PDA medium and preserved at 4°C. The deficient medium was used for preliminary screening of the soil. Selected molds were prepared into spore suspensions (106CFU/ml) by culturing fungi at 30°C in PDA medium and harvesting spores from two to three-day old cultures. Cultures were washed with 10ml of 0.5% sterile Tween 20 (R.T. Grade, Biodee Co. Ltd., Beijing) in distilled water (v/v). A suspension was then obtained by gentle shaking and filtering through three layers of sterile gauze to remove mycelia and sporangioconidia prior to centrifugation (TGL-16C, Anting Scientific Instrument Co. Ltd., Shanghai) at 302×g for 5min. The supernatant was discarded and the pellet was resuspended in 10ml of sterile 0.5% Tween 20. Spore densities were then measured using a hemacytometer and concentrations were used to adjust the final spore suspension to 106CFU/ml via dilution.
OTA solution (1.2ml of 10μg/ml stock) was added to 3.7ml of PDA medium along with 100μl of spore suspensions. Fungal cultures were incubated at 30°C for eight days, while bacterial cultures were incubated at 37°C for 2–3 days with shaking at 150rpm. Negative and positive controls were prepared by adding 1.2ml of OTA solution (10μg/ml) to either 3.8ml of sodium hypochlorite solution or media, respectively. After incubation, OTA was extracted with dichloromethane and measured using TLC to qualitatively analyze the OTA-degrading capacity of strains. Strains exhibiting OTA degradation activities were selected for additional analyses, and OTA degradation was further quantitatively analyzed by HPLC.
OTA and OTα detectionAfter incubation, degradation products were extracted after incubation with 5ml of dichloromethane, followed by drying of samples under a nitrogen gas flow. The resultant residue was again dissolved in 50μl of acetonitrile and 10μl of the mixture was spotted on silica gel plates (10×10cm, Qingdao Ocean Chemical Plant, Qingdao, China). The plates were then developed twice with ether and once with methylbenzene/ethylene/acetate/formic acid (6:3:1.4). OTA profiles were examined under UV light (365nm). To quantify OTA and OTα, 70% methanol culture extracts were collected in centrifuge tubes. The extractant solution was then evaporated off under a nitrogen gas flow, and 100μl of acetonitrile/water/acetic acid (99:99:2, v:v:v) was added. The extracts were then examined by HPLC (Agilent 1260, Santa Clara, CA, USA) using a C18 reversed-phase column (YMC-Pack ODS-AQ 250×4.6μm, 5mm) coupled to a fluorescence detector (λex=333nm; λem=460nm). The mobile phase was acetonitrile/water/acetic acid (99:99:2 v:v:v), flow speed was 0.8ml/min, and the column temperature was 35°C. Different concentrations (0.1, 2.5, 10, 20μg/l) of OTA standards were measured by HPLC, followed by quantifying peak areas. The signals of the samples were compared against the standard concentrations to determine the residual OTA concentrations after incubation. Specifically, OTA concentrations were calculated with a calibration curve exhibiting an R2=0.9998:
where X is the OTA concentration, and Y is the OTA peak area detected by HPLC.OTA concentrations were determined for both fungal cells and supernatant samples that were obtained by centrifugation at 302×g for 5min in order to ensure that the fungal strains could degrade OTA and that it was not simply adsorbed to the surface of cells. OTA was identified by its retention time (16.7min) based on that of a standard purchased from Sigma (Pribolab, Singapore). The OTα retention time was identified at 5.2min based on that of a standard prepared by total degradation of OTA by carboxypeptidase A (EC3.4.17.1) from bovine pancreas (Sigma, type II-PMSF). OTA and OTα concentrations were quantified by measuring sample peak areas and comparing against those of standard solutions. Residual OTA was thus calculated as follows:
where X1 is the OTA concentration in the negative control, X2 is the OTA concentration in the treated group, and Y is the degradation rate (%).Identification of fungal strainsIsolated strains that degraded OTA were identified based on morphological and culture characteristics31 in addition to 18S rDNA sequence analysis. The strains were first cultured on CA, CYA, and MEA media to observe culture characteristics via macro- and micro-morphological observations of mycelia and spores by microscopy and electron microscopy. Species identification was then performed by 18S rDNA sequence analysis.
Total genomic DNA was extracted from selected isolates after 72h of cultivation in malt extract medium (comprising 30g/l malt extract, 3g/l soy peptone, pH 5.6±0.2). Cells were then collected by centrifugation at 17000×g for 20min. Cells were then washed with distilled water and ground into powder for DNA extraction using a fungal DNA extraction kit. Extracted DNA was evaluated with 1% agarose gel electrophoresis to determine qualities and quantities.
The 18S rDNA gene of OTA-degrading isolates was amplified by PCR under standard conditions. The ITS1 (5′-TCCG TAGG TGAA CCTG CGG-3′) and ITS4 (5′-TCCT CCGC TTAT TGAT ATGC-3′) PCR primers were used for amplification35,36 and PCR products were visualized by electrophoresis. Phylogenetic analysis was conducted by aligning 18S rDNA consensus sequences that were generated at BGI Gene Technology Co., Ltd (Beijing, China) with references from the Genbank database that were identified by the BLAST analysis. A phylogenetic tree was constructed using neighbor joining methods in the MEGA 7.0 software package.
In addition, sequencing of β-tubulin and elongation factor (EF-1α) genes were used to confirm species identifications. Sequence similarities were expressed as the percentage of identity between the query sequences and the most closely related sequences in the databases. Strains were ultimately phylogenetically identified with phylogenetic analysis in the MEGA software.
OTA degradation product analysis and bioassays using strain M30011The molecular formulas of the OTA degradation products after treatment with strain M30011 were analyzed by High-Resolution Fourier Transform Ion Cyclotron Resonance Mass Spectrometry (HR-FT-ICR-MS) (Bruker Co. Ltd., Madison, USA). Stock solutions of OTA degradation products from strain M30011 were diluted with acetonitrile/water/acetic acid (99:99:2, v:v:v) to final concentrations of 10μM, and 20% methanol (methanol: water, v:v) and directly injected into the source region of the instrument at a rate of 4μl/min. All ESI mass spectra were acquired in the positive-ion or negative-ion mode by HR-FT-ICR-MS (Bruker Co. Ltd., Madison, USA).
Superoxide dismutase enzyme (SOD) activity and malondialdehyde (MDA) content were measured in Baby Hamster Syrian Kidney (BHK-21) cells. BHK-21 cells were cultivated at 37°C in a 5% CO2 atmosphere in Dulbecco's modified eagle medium (DMEM medium) supplemented with 10% bovine serum. Protein concentrations were measured using the Bradford method. Cellular SOD activity was measured using a cell SOD assay kit (Nanjing Jiancheng Bioengineering Institute, Nanjing, China). The MDA content in the culture medium was measured with an MDA assay kit (Nanjing Jiancheng Bioengineering Institute, Nanjing, China).
SOD activity was then calculated as follows:
where X1, X2, V1, V2, A, and Y are the OD of the control, the OD of the sample, the volume of the reaction system, the sample volume, the total protein content, and SOD activity, respectively. MDA content was calculated as follows:where X1, X2, Y1, Y2, A, D, and Z are the OD of the sample, the OD of the control, the OD of the standard, the OD of the standard control, the concentration of the standard sample, the diluted concentration, and the MDA content, respectively.Single-factor experiment to evaluate OTA detoxification by M30011The optimization of nitrogen nutrients (0.3% yeast extract) and carbon (3% lactose) nutrients in media has been previously carried out for strain M30011. Thus, additional factors influencing OTA degradation including media pH (5.0, 5.5, 6.0, 6.5, 7.0, 7.5, 8.0), incubation temperatures (25°C, 30°C, 35°C, 40°C, 45°C), and inoculum concentration (1×103, 1×104, 1×105, 1×106, 1×107, 1×108CFU/ml) were investigated.
Use of the response surface method (RSM) to optimize fermentation conditions for OTA detoxification by M30011Three key factors (temperature, inoculum concentration and pH) affecting the OTA degradation rate were selected for further analysis including temperature, inoculum concentration, and pH. A fractional factorial design was conducted to evaluate the influence of these factors on OTA degradation with four replications at the central point. The optimum conditions for enzyme activity were obtained by three primary analytical steps including the analysis of variance with ANOVA tests, regression analysis, and reaction surface modeling. First, the experimental results were analyzed by ANOVA, where p<0.05 was used to identify statistical significance. A regression coefficient was obtained by the multiple linear regression analysis, and a second-order polynomial model was used to fit the experimental data as follows:
where (i) a0, (ii) ai and aj, (iii) aii, ajj, and aij, and (iv) Xi and Xj are (i) the y-intercept, (ii) the linear coefficients, (iii) the interaction coefficients, and (iv) the coded levels of variables, respectively. RSM was used to determine the optimal degradation rate under different test conditions. Design-Expert 6.0 (Stat-Ease Inc., Minneapolis, MN, USA) was used to evaluate the fitness of the model via ANOVA and determination coefficients (R2 and adjusted R2). Finally, the optimal conditions for OTA degradation by strain M30011 were determined using the above information.Statistical analysesAnalysis of variance (ANOVA) and Duncan's multiple-range tests were performed to analyze the differences between experimental mean values. Values are shown as the mean of two experiments, where each condition was conducted triplicate.
Results and discussionScreening and isolation of OTA-degrading strainsThe mycotoxin OTA is found in agricultural and food products and one possible approach for removing it is via the application of an OTA degradation microbial strain. OTA has been quantified in agricultural and food products globally, and the maximum acceptable OTA levels have been identified by various organizations and countries7,8,19,21,40,43. Typical OTA levels in food products range between the detection limit and 35μg/kg. As indicated above, OTA can be biologically degraded in food processing. However, OTA levels, food processing conditions, and material properties are factors that influence detoxification treatments. Nevertheless, the safety of applying strains themselves is a critical issue that requires attention before widespread application.
The deficient solid medium with l-phenylalanine or isocoumarin supplemented as the only carbon source were used to identify strains with OTA degradation ability because the structures of l-phenylalanine and isocoumarin are similar to those of OTA. Thus, strains that can ferment l-phenylalanine or isocoumarin would also potentially degrade OTA. Hundreds of Chinese soil samples from different environments including cereal fields, forests, and gardens were used to screen for strains. A total of 130 bacterial and fungal strains that could metabolize l-phenylalanine and isocoumarin were isolated.
The ability of these strains to degrade OTA in liquid medium was then examined by TLC. Because OTA fluoresces under ultraviolet irradiation (365nm), the fluorescence of TLC plates could be used to further select strains that could degrade OTA. The OTA degradation ability of 130 isolates (80 bacterial and 50 fungal) were tested by TLC, yielding 25 strains that could degrade OTA (Table S1), indicating that those microorganisms could have excellent prospects for biodegradation of the mycotoxin. It is noteworthy that 19% of l-phenylalanine and isocoumarin metabolizing isolates did indeed exhibit OTA degradation ability. Thus, further screening was used to identify the possibility for higher degradation efficiencies among select microorganisms. Specifically, the production of extracellular OTA-degrading enzymes and associated activities were evaluated in the 21 bacterial and four fungal isolates with OTA degradation abilities.
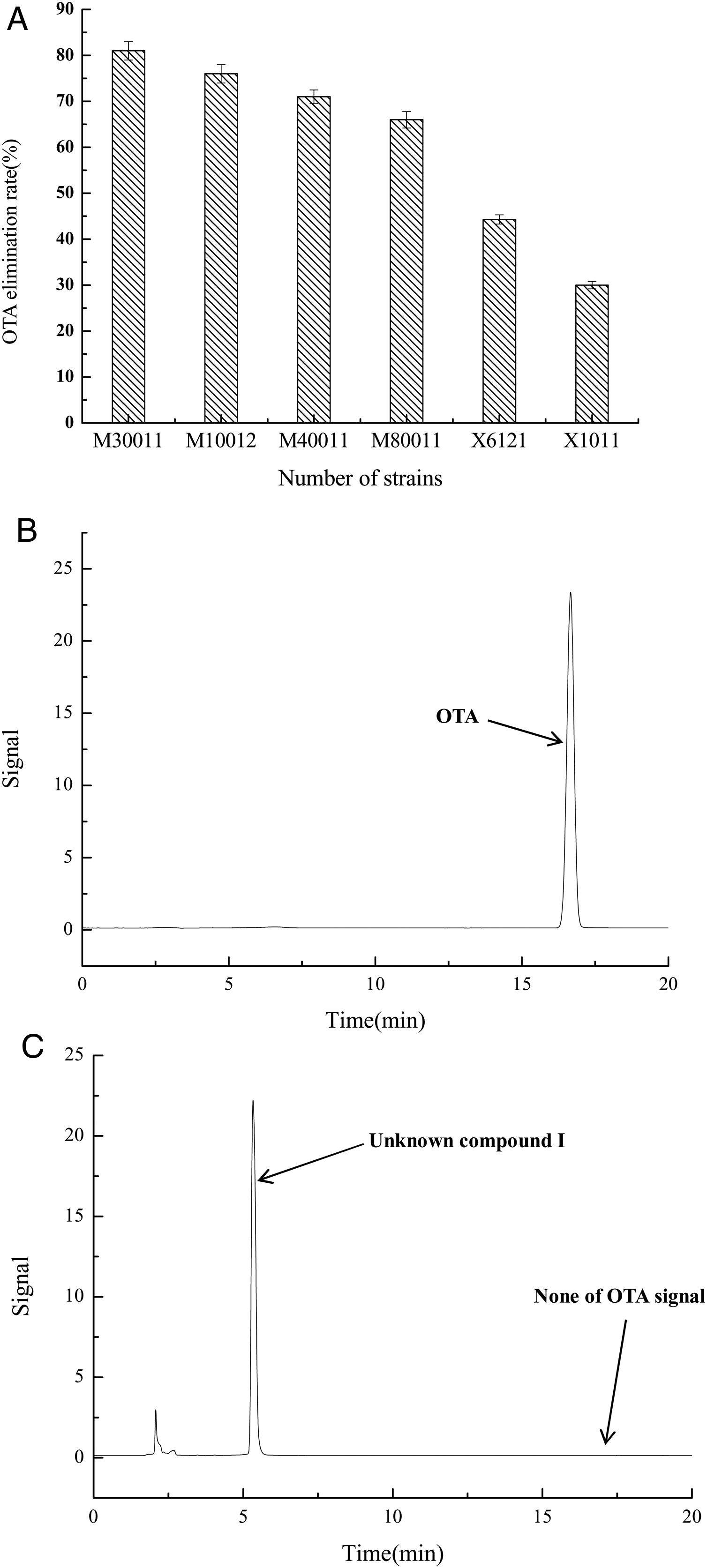
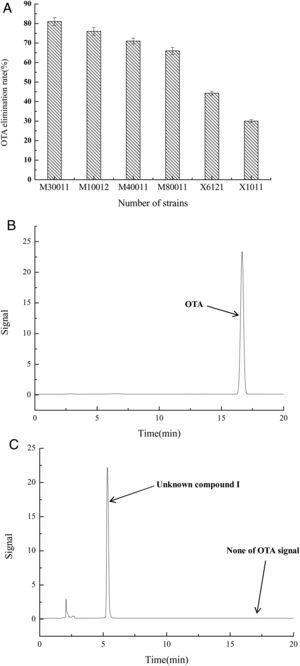
The OTA degradation rates of the isolated strains were measured by HPLC analysis. The OTA degradation rates in liquid medium for fungal strains M30011, M10012, M4001, and M80011 were 81%, 76%, 71%, and 66%, respectively (Fig. 1(A)). These rates were higher than those of the bacterial isolates (e.g., X1011, X6121, and other strains). Moreover, chromatograms revealed the reduction of OTA concentrations by the M30011 strain (retention time, 16.7min) and its conversion into an unknown compound, I (retention time, 5.2min) (Fig. 1(B) and (C)).
OTA degradation rate of strains was estimated based on HPLC. (A) OTA degradation rates of six strains. The relative OTA degradation rate was estimated based on HPLC chromatograms. (B) Chromatograms obtained for control group. (C) Chromatograms obtained for M30011 strain at pH 7.5 and 37°C, showing the hydrolysis of OTA. The retention time of OTA was 16.7min; the retention time of the unknown compound was 5.2min.
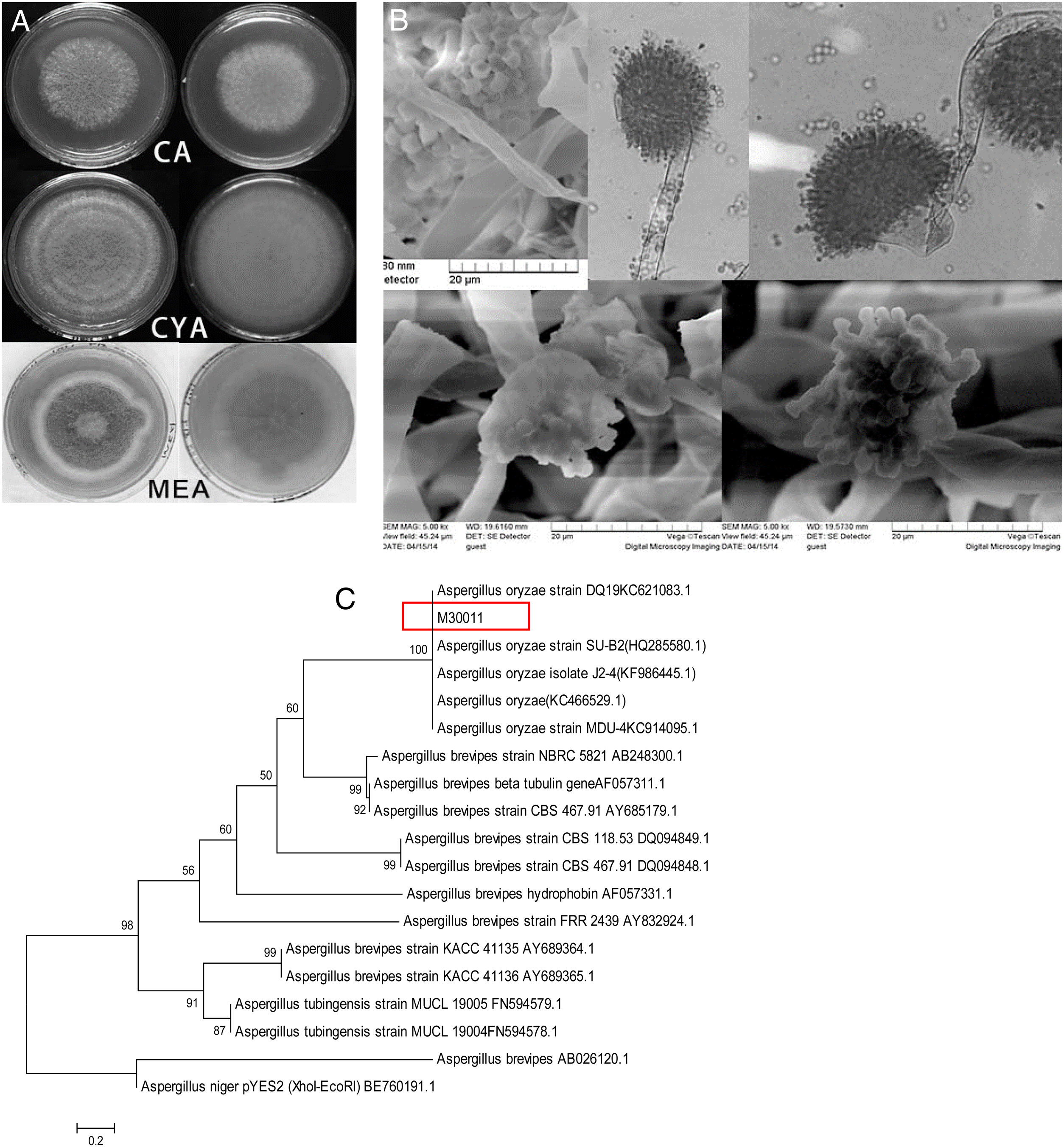
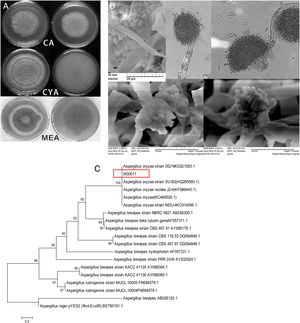
The four fungal strains were further identified as Aspergillus based on the 5th volume of the Flora Fungorum Sinicorum31. M30011 was selected for subsequent experiments because of its high OTA degradation ability. The isolate was further characterized by macro- and micro-morphological analyses (Fig. 2), in addition to its taxonomic classification using molecular analyses.
Identification of M30011 strain. (A) The growth morphology of M30011 on the CA, CYA and MEA medium. (B) Morphological characteristics of M30011 under microscope and electron microscope. (C) The phylogenetic tree of M30011 was constructed using near-full length 18S rRNA sequences by the neighbor-joining (NJ) and performed with 1000 replicate bootstrap resampling to form the overall consensus tree.
Phylogenetic analysis indicated that the 18S rDNA sequence of the strain was 100% identical to that of Aspergillus oryzae (Fig. 2(C)). The mycotoxin production capacity of strain M30011 was also investigated. The strain was not able to produce mycotoxins Aflatoxin B1 (AFB1), AFB2, AFG1, AFG2, OTA (data not shown) and cyclopiazonic acid (CA) (Fig. S1). Thus, these results all suggest that M30011 is closely related to A. oryzae.
Other strains exhibiting OTA degradation ability (M10012 and M40011) were also identified by sequence analysis (data not shown) and their 18S rDNA sequences shared 91% (M10012) and 99% (M40011) sequence identity to Aspergillus flavus. However, the safety risks associated with A. flavus unfortunately make their application in food industry difficult. In contrast, A. oryzae is a promising species for application in the food industry due to its lack of other health risks.
A. oryzae is a filamentous fungus that is often used in China, Japan, and Korea to ferment foods such as soybeans. It is also used to ferment rice and other cereals to make alcoholic beverages such as rice wine and sake in addition to vinegars such as rice vinegar. Indeed, A. oryzae has been used by humans as early as 2000 years ago33, and is considered a safe species for food products. Furthermore, when used in fermentation processes, A. oryzae can also reduce OTA levels in fermented products. Due to its importance, A. oryzae has been called a “national fungus” (Dr. Eiji Ichishima of Tohoku University, in the Journal of the Brewing Society of Japan) because of its importance in making koji, sake brewing, miso, soy sauce, and a range of other traditional foods. Thus, there is no safety risk associated with the application of A. oryzae in the food industry.
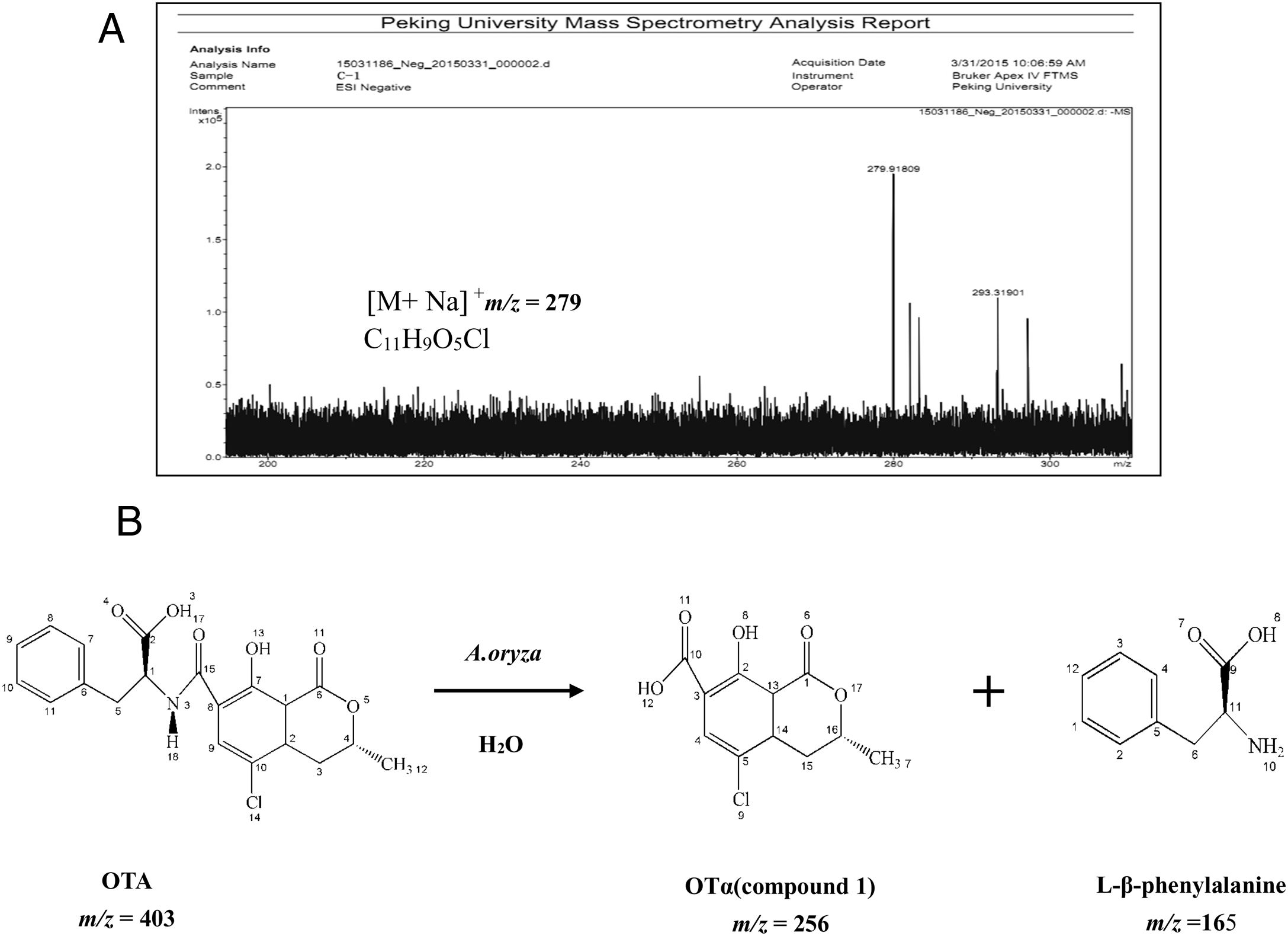
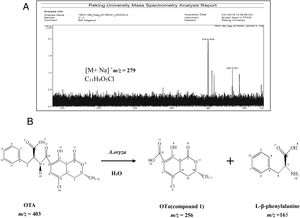
Analysis of OTA degradation products upon treatment with A. oryzaeThe molecular formula of the unknown OTA degradation product, compound I (Fig. 1(C)), was identified by HR-FT-ICR-MS (Fig. 3(A)). The sensitivity for detecting compound I was very low in the negative mode. In contrast, a [M+Na]+ ion peak at m/z 279 (relative intensity: 2.0×105) was obtained in the postive mode. Thus, compound I is positively charged based on the difference in sensitivity when detected in the positive and negative modes. After removing the sodium ion peak, the m/z was 256.9181, which corresponds to a molecular formula of C11H9O5Cl. Based on the chemical formula of compound I, we hypothesized that compound I was a hydrolysis product of OTA.
Numerous studies have shown that microorganisms can be used in OTA detoxification although analysis of several bacterial strains have indicated that most OTA detoxification can only be attributed to elimination (e.g., adsorption), rather than degradation. Moreover, some strains with degradation potential may also produce hazardous substances under certain conditions, leading to incomplete detoxification (degradation of OTA to safe products) and can lead to additional health risks.
The biotransformation of OTA in humans probably includes conversion of hydroxylated and/or dechlorinated cytochromes into P450 (CYP) products and secondary metabolites. These metabolites may especially include sugar esters (pentoses and hexoses) and phenylalanine compounds that have been detected in the culture medium of human hepatocytes. Both intestinal microflora and human cells metabolize OTA by cleavage of the peptide bond, producing the low-toxicity primary metabolite ochratoxin α (OTα, C11H9O5Cl)34. The primary product of OTA degradation by A. oryzae in a previous study was also OTα12. Accordingly, the retention time of the degradation product was about 5.3min (nearly the same as for OTα) under the same chromatographic conditions12. Collectively, these results indicate that the main OTA degradation product upon treatment with A. oryzae (C11H9O5Cl) was OTα. The detected concentration of OTα nearly corresponded to the theoretical concentration produced from the complete hydrolysis of OTA that was added to the medium (data not shown). The major pathway of detoxification of OTA by A. oryzae was similar to its biotransformation, resulting in OTα and phenylalanine (Fig. 3(B)). These results suggest the existence of a carboxypeptidase-like enzyme in A. oryzae that hydrolyzes the amide bond in the OTA molecule10,18,25.
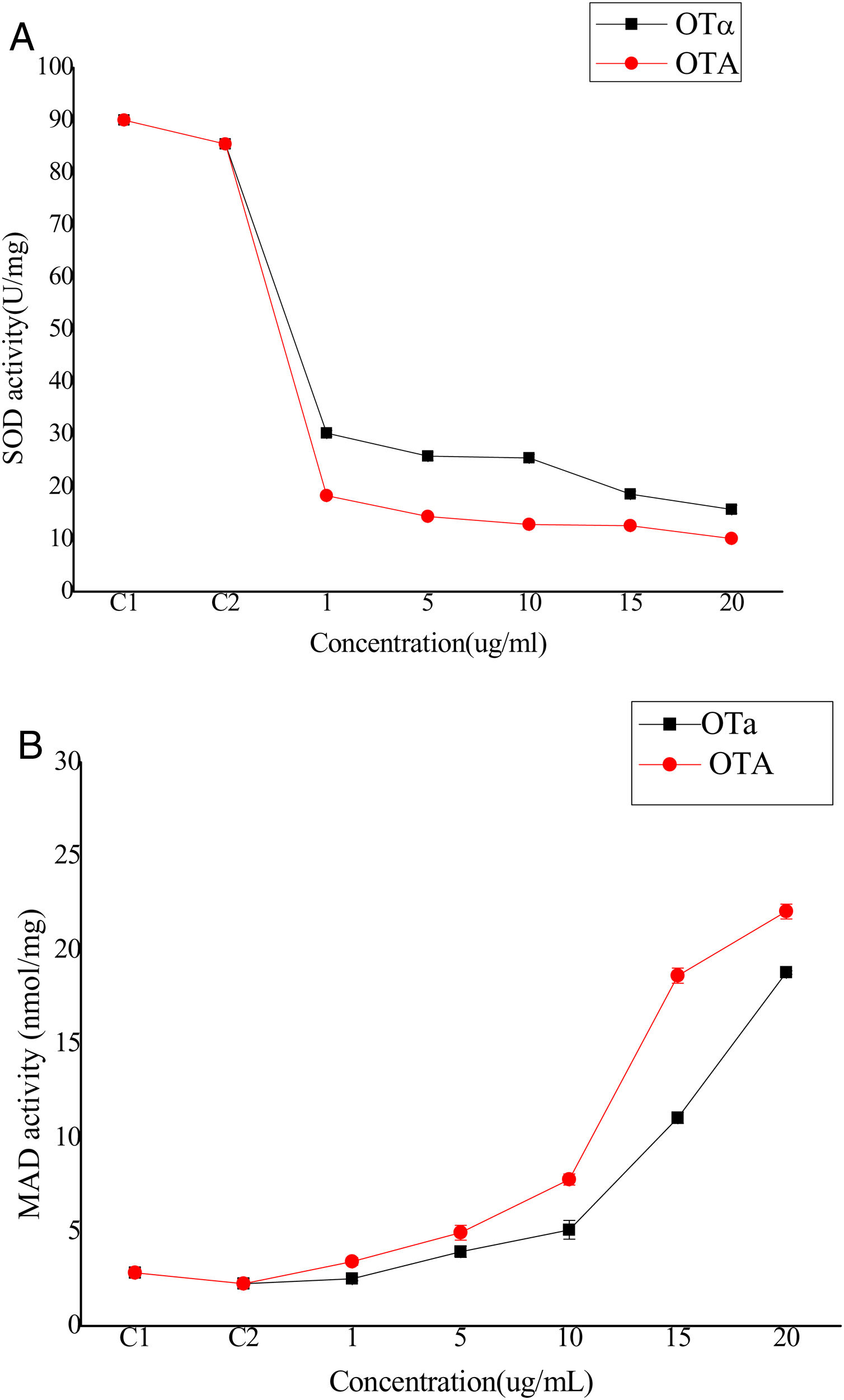
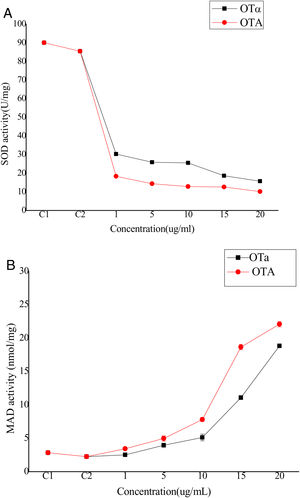
Bioassays of SOD activity and MDA content to evaluate biodetoxification capacityOTA exposure increases the intracellular production of reactive oxygen species (ROS) that cause oxidative damage to cells. Although the biological mechanisms by which OTA increases ROS production remain unclear, it has been suggested that ROS plays an important role in the toxicity of OTA to cells, possibly through lipid peroxidation and reduced antioxidant enzyme activity22. Thus, cell toxicity was evaluated by analyzing superoxide dismutase (SOD) activity and lipid peroxide content in cells treated with OTA or its main degradation product, OTα14,44. BHK-21 cells were treated with OTA or OTα for 24h at concentrations of 1, 5, 10, 15, or 20μg/ml, followed by the measurement of SOD activity. SOD activity decreased with increasing OTA concentrations (Fig. 4(A)). Specifically, the SOD activity of the cells treated with 1μg/ml OTA decreased to 21% (C2) (p<0.05) and 16%, 14%, and 11% for those treated with 5, 15, and 20μg/ml OTA, respectively. SOD activity was significantly higher after OTα treatment (at the same concentrations as above). For example, the SOD activity of cells treated with 1μg/ml OTα was 165% of that after OTA treatment. Likewise, the SOD activities of cells treated with 5 and 15μg/ml OTα was 180% and 150% of that after OTA treatment, respectively. Lastly, the SOD activity of cells treated with 20μg/ml OTα was 125% of that after OTA treatment. The decrease in SOD activity after treatment with 1μg/ml OTA was significantly higher than after treatment with 20μg/ml OTα (p<0.05).
The levels of the final lipid peroxidation product caused by ROS, MDA9, can reflect the degree of lipid peroxidation in the cell. MDA significantly affects the enzyme activity of mitochondrial respiratory chain complexes and indirectly reveals the degree of cellular damage. To evaluate potential lipid peroxidation, MDA content was measured in BHK-21 cells after treatment with OTA or OTα at various concentrations for 24h. MDA content increased with increasing OTA concentration treatments (Fig. 4(B)). Specifically, OTA treatments at concentrations of 1, 5, 15, and 20μg/ml resulted in increased MDA contents of 260%, 490%, 830%, and 980%, respectively, compared to controls. The increase in MDA content after OTα treatment was significantly lower than that obtained after OTA treatment. Specifically, OTα treatment at concentrations of 1, 5, 15, and 20μg/ml resulted in increased MDA contents of 110% (p>0.05), 175%, 227%, and 360%, respectively, compared to controls. The effects of 20μg/ml OTα treatment on MDA content were significantly weaker than those of 5μg/ml OTA (p<0.05).
Thus, these results indicate that OTA exposure significantly reduces SOD activity and increases MDA levels, thereby indicating strong effects on the activity of BHK-21 cells. After OTA treatment, intracellular lipid peroxidation increased, leading to increased intracellular lipid decomposition products and potential cellular metabolic dysfunction. In contrast, the cellular effects of OTα, the main OTA degradation product by A. oryzae, were significantly less severe, supporting the feasibility of using A. oryzae to detoxify OTA-contaminated agriculture and food products. Nevertheless, further animal toxicity studies are needed to confirm that A. oryzae treatment reduces OTA poisoning.
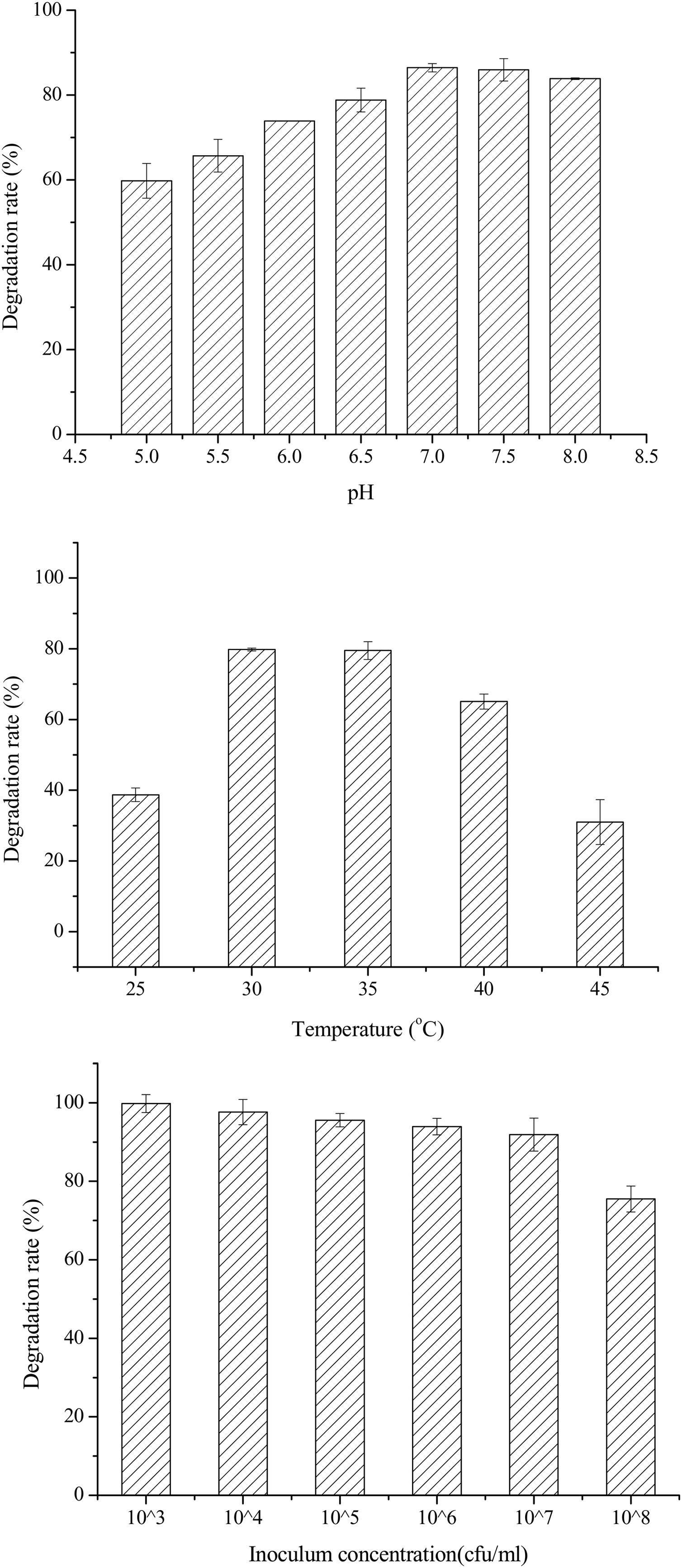
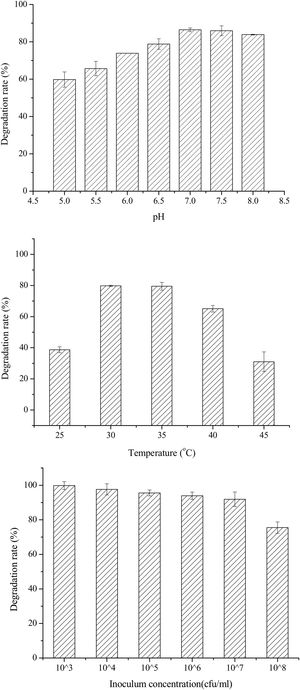
Single-factor optimization of OTA detoxification conditionsTo optimize conditions for OTA detoxification by A. oryzae, the influences of cultivation conditions on detoxification were individually investigated. The influence of pH on OTA degradation by A. oryzae was investigated in seven day fermentation experiments with medium containing 20g/l sucrose and 6g/l yeast extract (Fig. 5(A)). Over a pH range of 5.0–8.0, the maximum degradation rate (>86%) was observed at pH 7.0. Moreover, the OTA degradation rate strongly depended on pH, where degradation rates increased significantly as pH increased from 5.0 to 7.0. The OTA degradation rate was relatively high at a pH of 6.5 or 7.5 (78.81%–85.96%), while <84% of the OTA was degraded at pH 8.0. The OTA degradation rate also strongly depended on fermentation temperatures (Fig. 5(B)). At a fermentation temperature of 30°C or 35°C, the degradation rate was relatively high (79.80% and 79.50%, respectively), but less than 39% of the OTA was degraded at 25°C, and the degradation rate was only 31.0% at 45°C. Lastly, the inoculum concentration of A. oryzae was also a critical factor affecting OTA degradation (Fig. 5(C)). The maximum degradation rate (>99%) was observed at an inoculum concentration of 103CFU/ml when evaluated over a range of 103 to 108CFU/ml. Increased inoculum concentrations led to decreased OTA degradation rates. This observation, in addition to the preparation of higher inoculum concentrations requiring more medium, led to the consideration of 103CFU/ml as the optimal inoculum concentration for OTA degrading activity by A. oryzae.
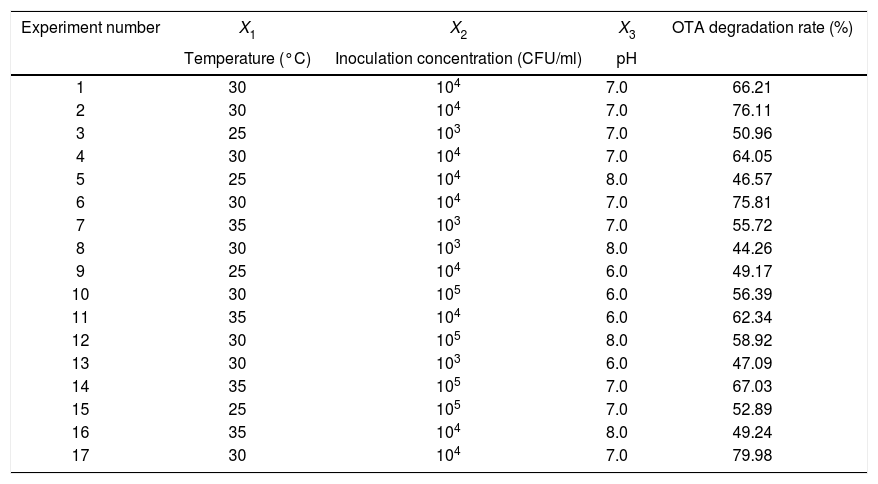
Response surface method (RSM) analysis to optimize the fermentation conditions for OTA detoxification by A. oryzaeBased on the results of the single-factor experiments, the following parameters were used to optimize the conditions for OTA detoxification by A. oryzae: pH, 6.0, 7.0, 8.0; temperature, 25°C, 30°C, 35°C; and inoculum concentration, 103, 104, 105CFU/ml. The design and results of the Box–Behnken optimization experiment are shown in Table 1. Quadratic multinomial regression fitting was conducted to identify the response (Y) value (i.e., the OTA degradation rate) for pH, temperature, and inoculum concentration conditions, as calculated with the following quadratic multinomial regression equation:
Design and results of Box–Behnken optimization experiments.
| Experiment number | X1 | X2 | X3 | OTA degradation rate (%) |
|---|---|---|---|---|
| Temperature (°C) | Inoculation concentration (CFU/ml) | pH | ||
| 1 | 30 | 104 | 7.0 | 66.21 |
| 2 | 30 | 104 | 7.0 | 76.11 |
| 3 | 25 | 103 | 7.0 | 50.96 |
| 4 | 30 | 104 | 7.0 | 64.05 |
| 5 | 25 | 104 | 8.0 | 46.57 |
| 6 | 30 | 104 | 7.0 | 75.81 |
| 7 | 35 | 103 | 7.0 | 55.72 |
| 8 | 30 | 103 | 8.0 | 44.26 |
| 9 | 25 | 104 | 6.0 | 49.17 |
| 10 | 30 | 105 | 6.0 | 56.39 |
| 11 | 35 | 104 | 6.0 | 62.34 |
| 12 | 30 | 105 | 8.0 | 58.92 |
| 13 | 30 | 103 | 6.0 | 47.09 |
| 14 | 35 | 105 | 7.0 | 67.03 |
| 15 | 25 | 105 | 7.0 | 52.89 |
| 16 | 35 | 104 | 8.0 | 49.24 |
| 17 | 30 | 104 | 7.0 | 79.98 |
The model exhibited high reliability and good statistical performance, with a regression probability of 95%, and no lack of fitting. The high correlation coefficient (R2) of 0.882 indicated that the model explained 88.2% of the response variability. Further, the response percentage and high F-value (5.79) predicted by the model indicated that the model fit was indeed very significant.
The interactions among various factors of the response surface and the contour plot are shown in Figure S1. Temperature and inoculum concentrations both significantly influenced OTA degradation, when considered independently or simultaneously (Fig. S1(A)). Based on the model, the degradation rate was highest when using 104CFU/ml and an incubation temperature of 30°C. Using the highest inoculum concentration led to a low degradation rate, possibly due to the scarcity of oxygen and nutrients. Further, when both parameters were set to optimal conditions, the model did not indicate the presence of any additional positive effects. Considering the level of influence of each single parameter (Fig. 5(B) and (C)), no statistical difference was observed between the predictions and the results using parameter combinations (Fig. S2(A)). When the lowest or highest temperatures were combined with high or low inoculum concentrations, low degradation rates were consistently obtained. In all of these cases, the degradation rates were much lower than expected from the single parameter experiments. Thus, these results indicate the importance of selecting the right combination of temperature and inoculum concentrations to achieve optimal OTA degradation.
Temperature and pH combined did not significantly affect the OTA degradation rate (Fig. S2(B)). Moreover, pH and inoculum concentrations combined did not significantly impact the OTA degradation rate (Fig. S2(C)). Consequently, the response surface optimization model predicted that the optimum pH, temperature, and inoculum concentrations to use for cultivation were 8.0, 30°C, and 104CFU/ml. The model was verified by parallel experiments, followed by OTA content measurement by HPLC. The maximum degradation rate observed was 94% after 72h. Thus, the time necessary for OTA degradation has decreased by 57% after optimization of experimental conditions. These results further support the potential for applying A. oryzae in agricultural and food industry settings for OTA degradation.
FundingAll supported research funding is indicated in the Acknowledgements.
Conflict of interestThe authors declare that they have no conflicts of interest.
Informed consentInformed consent was obtained from all of the authors and the authors agreed to the submission of the manuscript to the journal.
This study was funded by the National Key R&D Program of China (No. 2017YFC1600605), the National Natural Science Foundation (No. 31601408), the Beijing Natural Science Foundation (No. 6172003), and the Scientific & Technological Innovation Service Capability Construction Project (No. PXM2018_014213_000033).