Campylobacter jejuni is an important foodborne pathogen with global distribution. We describe a genotyping study of a collection of C. jejuni (n=137) isolated from different broiler farms and from multiple sites along the processing line in a slaughterhouse in Argentina during 2011, 2012 and 2015. The isolates were genotyped using pulsed-field gel electrophoresis (PFGE) and multilocus sequence typing (MLST). Based on the PFGE results, the isolates were grouped into 26 pulsotypes. Subsequently, the isolates representing these 26 pulsotypes were chosen for MLST genotyping, which identified 16 different sequence types (STs) and 6 clonal complexes (CCs) (21, 45, 48, 353, 354, 446). Several of the STs (n=7) have not been previously reported in the PubMLST.org database. The most prevalent CCs were 21, 45 (both associated with human campylobacteriosis worldwide) and 353. This study showed high genetic diversity among C. jejuni in the broiler production environment in Argentina with novel MLST genotypes.
Campylobacter jejuni es un importante agente patógeno de transmisión alimentaria a nivel mundial. Describimos el estudio molecular de una colección de C. jejuni (n = 137) aislados de diferentes granjas de pollos y de múltiples sitios de un frigorífico de Argentina durante los años 2011, 2012 y 2015. Los aislamientos fueron genotipados mediante electroforesis en campo pulsado (PFGE) y tipificación multilocus de secuencias (MLST). Con base en los resultados de PFGE, los aislamientos se agruparon en 26 pulsotipos. Se seleccionaron aislamientos representativos de dichos pulsotipos para genotipificarlos mediante MLST, se identificaron así 16 secuenciotipos (ST) diferentes y seis complejos clonales (CC) (21, 45, 48, 353, 354, 446). Varios de los ST (n = 7) no habían sido reportados previamente en la base de datos PubMLST.org. Los CC más prevalentes fueron el 21, el 45 (asociados con casos de campilobacteriosis) y el 353. Este estudio identificó una elevada diversidad genética de C. jejuni en la cadena cárnica aviar argentina con novedosos genotipos mediante MLST.
Thermotolerant Campylobacter, specifically Campylobacter jejuni is a leading cause of zoonotic enteric infection in both developed and developing nations4. Broiler meat is one of the most important sources of human campylobacteriosis (EFSA, 2019)5. For surveillance and epidemiological studies, molecular typing is a commonly used tool to analyze Campylobacter isolates derived from various sources. Among the various molecular typing methods used for C. jejuni, pulsed-field gel electrophoresis (PFGE) combined with multilocus sequence typing (MLST) is considered the gold standard due to its high discrimination potential7. Another advantage of MLST is that data are stored in an internet-based database, facilitating interlaboratory comparisons3. Although whole genome sequencing is the best typing tool in terms of sensitivity and accuracy, it is more expensive and may not be practical for resource-limited laboratories.
Molecular typing is very important for understanding the epidemiology of C. jejuni at the regional, national and international levels. Genotyping has shown that C. jejuni from different sources is genetically diverse and a large number of strain types exist regardless of the genotyping methods used8. Despite the importance of C. jejuni in foodborne disease, little is known about its distribution and genotypes in the broiler production system in Argentina. This study was conducted with the aim of evaluating genetic diversity among thermotolerant Campylobacter isolated from Argentina and comparing our results with the MLST profiles of C. jejuni isolates from South America. In the present study, we used PFGE and MLST to investigate which C. jejuni genotypes were circulating in the broiler meat chain in Argentina. A stratified approach was used, in which a large number of isolates were first typed by PFGE, and then a subset of the C. jejuni isolates was selected for analysis by MLST. In addition, we compared our results with published information from South America. To the best of our knowledge, this is the first report of C. jejuni MLST profiles in Argentina.
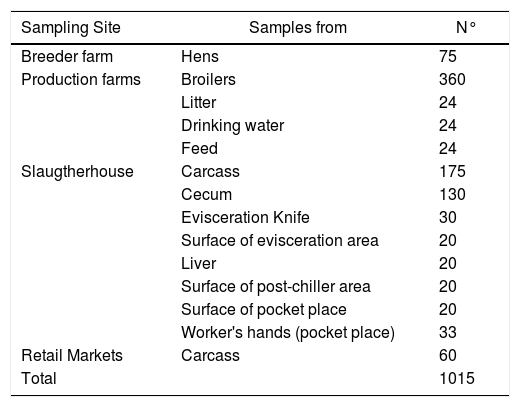
Sample collection and Campylobacter isolationDifferent samples were taken from broiler farms (breeder and production farms), a slaughterhouse and retail markets from Santa Fe province in Argentina during the years 2011, 201216, and 2015 (Table 1).
Source and number of samples.
| Sampling Site | Samples from | N° |
|---|---|---|
| Breeder farm | Hens | 75 |
| Production farms | Broilers | 360 |
| Litter | 24 | |
| Drinking water | 24 | |
| Feed | 24 | |
| Slaugtherhouse | Carcass | 175 |
| Cecum | 130 | |
| Evisceration Knife | 30 | |
| Surface of evisceration area | 20 | |
| Liver | 20 | |
| Surface of post-chiller area | 20 | |
| Surface of pocket place | 20 | |
| Worker's hands (pocket place) | 33 | |
| Retail Markets | Carcass | 60 |
| Total | 1015 |
Fecal samples (hens from breeder flocks and broilers) were randomly collected from the cloaca using sterile cotton swabs, which were placed in capped plastic tubes containing 10ml of Cary-Blair (Britania®) transport medium and transported to the laboratory under refrigeration conditions within 4h. Together with the cloacal samples, samples of broiler feed (500g), drinking water (1 l) and litter (500g) were also taken from each flock. Feed, drinking water, and litter samples were taken directly from the feeders. Cecal and liver samples in the slaughterhouse were randomly collected from the evisceration line and placed into sterile plastic bags. Broiler carcasses were taken from the processing line after chilling, using a clean pair of latex gloves and put into a sterile bag with 200ml of Ringer's solution ¼ strength. Carcasses were rinsed by shaking for 60seconds in each of two directions to ensure that the solution came into contact with all the surfaces; the solution was recovered and transported to the lab in sterile plastic tubes (under refrigeration conditions) within 4h. Samples were taken from the slaughterhouse surfaces, evisceration knives, and workers’ hands using sterile cotton swabs which were placed in capped plastic tubes containing 10ml of Cary-Blair and transported to the laboratory under refrigeration conditions within 4h. Broilers were packaged at the processing plant in the slaughterhouse and transported to the retail market where they were randomly sampled, following the same procedure described for the broiler carcasses in the slaughterhouse. All the samples were placed in selective enrichment broth in a 1:9 proportion (sample:broth)
Campylobacter isolates were obtained using the selective enrichment media Bolton Broth and Preston Agar1. All incubations were performed under microaerophilic conditions (5% O2, 10% CO2 and 85% H2) at 42°C. Positive isolates were subcultured on Columbia blood agar and stored in glycerol broth (15% glycerol and 85% serum broth) at−80°C.
DNA extraction and identification of C. jejuniDNA was extracted using the Wizard® Genomic DNA Purification Kit (Promega). All presumptive Campylobacter spp. isolates were identified as C. jejuni by PCR, as described by Vandamme et al.12
PFGE-typingThe analysis of C. jejuni isolates by PFGE was performed according to the method described in the PulseNet protocol using SmaI (Fermentas®) as the restriction endonuclease. Salmonella spp. H9812 was used as reference marker (digested with Xba I; Fermentas®). PFGE banding patterns were analyzed using BioNumerics version 6.6 (Applied Maths, Belgium). Images of gels were normalized by alignment with the appropriate size markers. Matching and dendrogram of fingerprints were determined by the unweighted pair group method with averages (UPGMA) and performed using the Dice coefficient (position tolerance, 1.0%). The PFGE cluster was based on a 95% similarity cut-off.
MLST-typingMultilocus sequence typing (MLST) of C. jejuni isolates was performed as described elsewhere3. Briefly, PCR was used to amplify a segment of seven housekeeping genes: aspA, glnA, gltA, glyA, pgm, tkt, and uncA. PCR and sequencing reactions were carried out according to the guidelines provided on the Campylobacter MLST website (https://pubmlst.org/campylobacter/info/). Each PCR amplification mixture contained: 50 ng genomic DNA, 1×MasterAmp PCR buffer (Takara, Japan), 2.5mM MgCl2, 250μM (each) dNTPs, 50pmol of each primer, and 1 U Ex-Taq polymerase (Takara, Japan). Amplicons were purified using the QIAquick PCR Purification Kit (Qiagen, Hilden, Germany) and sequenced by the DNA Facility of the Iowa State University Office of Biotechnology using the 3730xl DNA Analyzer (Applied Biosystems, Auckland, New Zealand). Allele numbers and sequence types (STs) were assigned using the Campylobacter MLST website (developed by Keith Jolley and sited at the University of Oxford; http://pubmlst.org/campylobacter/). Novel alleles and STs were submitted to the Campylobacter MLST website curators for number assignment.
In total, 137C. jejuni isolates were typed by PFGE. Overall, the PFGE analysis indicated the presence of 26 clusters comprising four or more isolates along with 81 unique genotypes. One cluster contained 12 isolates from samples from farms (fecal samples from broilers) and the slaughterhouse (carcass rinse). Many of the clusters contained just a few isolates (two or three). These results revealed a very high degree of genetic heterogeneity among the C. jejuni isolates.
In order to further define the genotypes, 26C. jejuni (representing different PFGE clusters) were chosen for MLST typing. MLST characterization of the 26 isolates yielded 9 previously known STs and 7 new STs. The STs detected include ST-50, ST-137, ST-446, ST-475, ST-2109, ST-6091, ST-6669, ST-8498, ST-8499 and the most prevalent ST was ST-137. These STs were isolated from samples taken in farms, a slaughterhouse and retail markets. The new STs assigned by the curators were ST-8500, ST-8501, ST-8502, ST-8503, ST-8504, ST-8505, ST-8506, ST-8507, being isolates from breeder and production farms.
With regard to the clonal complex (CC), six CC groups were identified and the most prevalent, in descending order, were CC-45(n=10), CC-21 (n=4), CC-353 (n=3), CC-354 (n=1), CC-446 (n=1) and CC-48 (n=1). It was impossible to assign six isolates to a CC. CC-45 alone accounted for 42% of the isolates and together with CC-21 accounted for 61% of the whole dataset. Additionally, CC-45 was detected in the slaughterhouse samples and retail markets, CC-21 in samples from farms, the slaughterhouse and retail markets, and CC-353 in farms and the slaughterhouse. Clonal complexes 354 and 446 were detected only in farms and CC-48 was detected in the slaughterhouse.
C. jejuni is a multihost and genetically diverse pathogen and therefore discriminatory genetic typing methods are needed for source attribution and epidemiological investigation. For this purpose, PFGE and MLST have been widely used for Campylobacter typing and resolving the relationship of genetically closely related bacterial isolates10.
The results of our study showed that the different stages of the broiler meat production chain exhibited different C. jejuni PFGE profiles. Several reports showed a similar number of clusters for C. jejuni and many unique profiles for this species10. The genetic diversity of Campylobacter was also reported in previous studies14. Genetic diversity can be generated by the acquisition of foreign DNA, spontaneous mutation, or recombination of large DNA segments, all of which could alter the PFGE patterns13.
Additionally, this study determined sequence types and clonal complexes of the isolates by MLST. Nine STs were found among the analyzed strains. It is important to mention that if more profiles of PFGE had been analyzed, especially those representing a unique profile, a greater genetic diversity would have been detected. Furthermore, five clonal complexes (CC-45, CC-21, CC-353, CC-354 and CC-446) were found among our strains. This is in agreement with different studies demonstrating those genotypes as dominant among the broiler and human C. jejuni population in various geographic regions2. According to the PubMLST.org database, CC-21 is considered one of the most prevalent and widely distributed genotypes among human clinical strains15. Elsewhere, CC-45 is also a major CC frequently isolated from human clinical samples in European studies15.
Moreover, six of the 26 isolates could not be assigned to any known clonal complexes. This level of genetic variation is similar to what was observed in other studies of broiler materials, particularly since broiler isolates have shown higher diversity than, for instance, human isolates9. This variation may reflect the differences in production systems, animal health, and climatic conditions, among other potential variables among countries. These factors could affect the epidemiology of Campylobacter and the implication of the new profiles in the epidemiology of Campylobacter needs further research.
Most of the MLST data present in the Campylobacter database (http://pubmlst.org/ campylobacter/) correspond to strains isolated in countries from Europe, Oceania and North America. A query in the C. jejuni PubMLST.org database (Last accessed: 07/12/2018) showed that only Brazil, Uruguay, Ecuador, Chile and now Argentina have reported C. jejuni MLST profiles from human campylobacteriosis and MLST profiles from broiler isolates. The information is important for understanding the epidemiology and should be considered as the scientific basis to implement risk management measures to protect public health.
In addition, CC-45, CC-21 and CC353 grouped 73% of the isolates from the broiler production chain in Argentina. When compared with CCs in South America (PubMLST.org database), we found that CC-353 is the most prevalent in Brazil, while CC-21 followed by CC-353 is dominant in Chile (PubMLST.org database2). These similarities in CCs among countries in the South American region may be explained by commercial relationships among the nations as export and import animal food products are very common and animal livestock production is one of the most common points in the commercial relationship. This could explain why the 2017 official record indicated that Argentina imported meat broilers from Brazil (90% of the total imports) and exported 9% of its production to Chile11. Different results were found in Uruguay, where eleven isolates grouped into 9 CCs and did not show a prevalent CC, which might be due to the limited number of isolates in the database. Thus, information from Uruguay cannot be validly compared with that from other South American countries.
PubMLST.org database showed that CC-353, CC-21 and CC-48 are the most prevalent CCs related to isolates from clinical human cases in South America. Consistently with the notion that broilers are a source of campylobacteriosis, we found CC-353, CC443 (only reported by Brazil), CC 45 (only reported by Argentina) and CC21 reported by both countries. This relationship between CCs from human cases and broilers further support that broiler meat serves as a route of infection of Campylobacter in humans.
The most prevalent CCs found in human cases are CC-21, CC-828 and CC-48, while in broiler sources are CC-828, CC-21 and CC-45. When we compare the information with MLST data from South America, we found differences among the most prevalent CCs. Earlier studies have shown that host specificity of genotypes overrides the geographical location6, meaning that there are several genotype assemblages associated with different niches with large spatial distributions7. This geographic distribution could be, in part, a possible explanation for the differences between South America and the rest of the world.
The present work is the first molecular typing study of Campylobacter in the broiler production chain in Argentina, revealing that C. jejuni showed high genetic diversity by PFGE and MLST typing. It is clear that broilers are colonized on the farm by diverse strains of C. jejuni; therefore, further studies should be conducted to evaluate the importance of different vehicles for the transmission of this foodborne pathogen. This collective evidence should be considered the scientific basis to implement risk management measures to protect public health.
FundingsThis study was funded by PICT-Joven N°1491/2014 (Agencia Nacional De Promoción Científica Y Tecnológica).
Conflict of interestNone.
The authors would like to thank Dr. Gerardo Leotta (Instituto de Genética Veterinaria (IGEVET-CONICET/UNLP) Facultad de Ciencias Veterinarias (UNLP) for the use of the Bionumerics software. We also thank Qijing Zhang for his assistance and cooperation in the training of our working group in molecular techniques of thermotolerant Campylobacter.