Different phenotype-based techniques and molecular tools were used to describe the distribution of different Achromobacter species in patients with cystic fibrosis (CF) in Argentina, and to evaluate their antibiotic resistance profile. Phenotypic identification was performed by conventional biochemical tests, commercial galleries and MALDI-TOF MS. Genetic approaches included the detection of A. xylosoxidans specific marker blaoxa-114, the amplification and sequencing of the 16S rRNA gene, nrdA and blaOXA complete sequence, and MLST analysis. Phenotypic approaches, even MALDI-TOF, rendered inconclusive or misleading results. On the contrary, concordant results were achieved with the nrdA sequencing or sequence type (ST) analysis, and the complete blaOXA sequencing, allowing a reliable discrimination of different Achromobacter species. A. xylosoxidans accounted for 63% of Achromobacter infections and A. ruhlandii accounted for 17%. The remaining species corresponded to A. insuavis, A. dolens, A. marplatensis and A. pulmonis. Antimicrobial susceptibilities were determined by the agar dilution method according to CLSI guidelines. Piperacillin, piperacillin/tazobactam and carbapenems were the most active antibiotics. However, the emergence of carbapenem-resistant isolates was detected. In conclusion, prompt and accurate identification tools were necessary to determine that different Achromobacter species may colonize/infect the airways of patients with CF. Moreover, antimicrobial therapy should be administered based on the susceptibility profile of individual Achromobacter sp. isolates.
Se emplearon diversas técnicas fenotípicas y moleculares para describir la distribución de diferentes especies del género Achromobacter en pacientes con fibrosis quística (FQ) en Argentina, y se evaluó el perfil de resistencia a los antibióticos. Se realizó la identificación fenotípica por pruebas bioquímicas convencionales, galerías comerciales y MALDI-TOF MS. El enfoque genético incluyó la detección del marcador especie-específico de A. xylosoxidans blaoxa-114, la amplificación y la secuenciación de los genes ARNr 16S, nrdA y secuencia completa de blaOXA, y el análisis por MLST. Los enfoques fenotípicos, incluso la técnica de MALDI-TOF, proporcionaron resultados no concluyentes o erróneos. Por el contrario, se obtuvieron resultados concordantes entre la secuenciación del gen nrdA o el análisis de secuenciotipos (ST) y la secuenciación completa de blaOXA, lo que permitió una discriminación confiable de las diferentes especies de Achromobacter. A. xylosoxidans representó el 63% de las infecciones por Achromobacter y A. ruhlandii representó el 17%. Las especies restantes correspondieron a A. insuavis, A. dolens, A. marplatensis y A. pulmonis. Se determinó la sensibilidad a antimicrobianos por el método de dilución en agar de acuerdo al CLSI. Los antibióticos más activos fueron piperacilina, piperacilina/tazobactam y carbapenemes. Sin embargo, se detectó la emergencia de aislamientos resistentes a carbapenemes. En conclusión, resultaron necesarias herramientas de identificación rápida y precisas para determinar las diferentes especies del género Achromobacter capaces de colonizar/infectar las vías respiratorias de los pacientes con FQ. Asimismo, la terapia antimicrobiana debería llevarse a cabo en función del perfil de sensibilidad de los aislamientos individuales de Achromobacter spp.
Achromobacter spp. are increasingly recognized as emerging pathogens in patients with cystic fibrosis (CF)20. Reported rates of Achromobacter colonization/infection in individuals with CF, vary between 2% and 17.9%, and display a rising tendency worldwide3,10. Achromobacter xylosoxidans is the most frequent species recovered within this genus, however other species have been associated with human infections15,17. Moreover, clinical isolates are mostly referred as A. xylosoxidans given that the accurate species identification of Achromobacter isolates is difficult.
Conventional phenotypic methods have been commonly used for bacterial identification of Achromobacter spp. in many clinical laboratories, as their implementation and cost make them more affordable. Since these classical methods yield unclear results, and generally fail to differentiate between species of the genus, molecular methods have been proposed as complementary or alternative procedures.
Amplification and sequencing of the 16S rRNA coding gene, which constitutes a useful method in the identification of numerous microorganisms, is not able to discriminate between species of Achromobacter5,12. In 2011, Turton et al. proposed the amplification of the blaOXA-114 gene for rapid and accurate A. xylosoxidans identification9,21. Moreover, in 2013 we described the presence of blaOXA-258, blaOXA-364, and blaOXA-243 in Achromobacter ruhlandii, Achromobacter dolens, and Achromobacter insuavis, respectively, as species-specific markers useful for bacterial identification14,19. Simultaneously, Spilker et al. proposed the amplification and sequencing of an inner fragment of the nrdA gene, one of those included in the multilocus sequence typing (MLST) scheme, as a precise method for the identification of different species of Achromobacter17.
MALDI-TOF MS has emerged as a revolutionary technique for rapid bacterial identification. This method has been shown to be more rapid, accurate and cost-efficient than conventional phenotypic techniques or genotypic approaches. However, its performance is uncertain in infrequent species.
In this study, different phenotype-based techniques and molecular tools were conducted to describe the distribution of different species of Achromobacter in patients with CF in Argentina, and to evaluate their antibiotic resistance profile.
MethodsIsolatesForty-one non-related Achromobacter spp. clinical isolates, recovered from patients with CF at 6 healthcare centers in Argentina during 1996–2013 were included. Isolates were mainly obtained from respiratory secretions (Table S1, supplementary material).
Phenotypic identificationPhenotypic identification was performed by biochemical tests according to Yabuuchi et al.23 and Vandamme et al.22 The biochemically-based commercial system, API 20NE (bioMèrieux) was also conducted.
MALDI-TOF MS identification was performed using a Microflex MALDI-OF MS instrument (Bruker Daltonics, GmbH, Germany) and FlexControl 3.0 software (Bruker Daltonics). Identification scores ≥2.0 were accepted for a reliable identification at species level and scores between ≥1.7 and ≤2.0 were accepted for identification at genus level. Scores <1.7 indicated no reliable identification8.
Genotypic identificationThe 16S rRNA gene was amplified by PCR as previously described13. Purified amplicons were sequenced in both strands using an ABI Prism DNA 3700 sequencer and compared with databases using the NCBI's BLAST tool.
The nrdA sequence was achieved according to Spilker et al. and compared with databases (https://pubmlst.org/achromobacter/)17.
The presence of A. xylosoxidans species-specific marker, blaOXA-114, was investigated by PCR amplification as previously described by Turton et al.21 The complete blaOXA-114, blaOXA-258, blaOXA-364 and blaOXA-243 sequences intrinsic for A. xylosoxidans, A. ruhlandii, A. dolens, and A. insuavis, respectively, were amplified according to Traglia et al.19 Amplicon sequences were compared with those of all the different blaOXA variants deposited in GenBank.
Multilocus sequence typing analysisA multilocus sequence typing (MLST) scheme was conducted to identify those isolates which could not be unambiguously identified by nrdA sequencing. For this purpose, amplification and sequencing of inner fragments of seven housekeeping genes were performed and the corresponding allele profiles and sequence types (ST) were assigned according to the Achromobacter MLST website (http://pubmlst.org/achromobacter/)18.
Antimicrobial susceptibility testingMinimal inhibitory concentrations (MIC) were determined for a representative set of antibiotics: ampicillin, piperacillin, cefoxitin, ceftazidime, cefepime, piperacillin/tazobactam, imipenem, meropenem, ciprofloxacin, levofloxacin, amikacin, kanamycin, gentamicin, trimethoprim-sulfamethoxazole, tetracycline and colistin. Antimicrobial susceptibilities were determined by the agar dilution method according to Clinical and Laboratory Standards Institute (CLSI) recommendations. The susceptibility breakpoints used in this study were those established for other non-Enterobacteriaceae6.
ResultsPhenotypic identification results are shown in Table S1. According to classical biochemical methods, 27/41 isolates were identified as A. xylosoxidans, the remaining ones being identified as Achromobacter spp. Using the API 20NE commercial gallery, 39/41 corresponded to A. xylosoxidans. Based on MALDI-TOF MS data, all isolates except one were consistent with A. xylosoxidans. Species identification was achieved in 25/40 isolates with score ≥2, while 15/40 displayed scores from 1.7 to 1.9. None of the samples presented score <1.7.
The 16S rRNA sequences obtained for the isolates included in this study displayed about 99% identity with those deposited for different species of Achromobacter, being unable to discriminate among them.
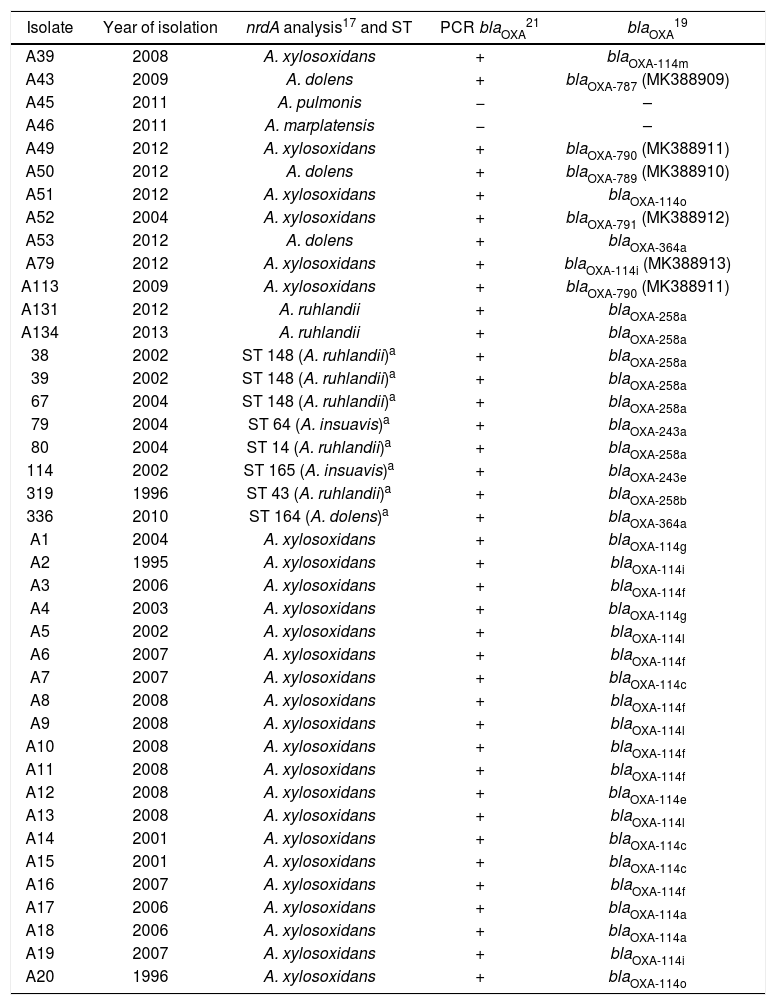
The results of the other genetic approaches performed in this study are shown in Table 1. The amplification of blaOXA-114 proposed by Turton et al.21 yielded positive results in 39/41 isolates, indicating that they corresponded to A xylosoxidans. However discrepancies were observed in 13/39 isolates when comparing these results with those obtained using other molecular methods. Based on these methods, the 13 isolates were finally identified as Achromobacter species other than A. xylosoxidans (Table 1).
Comparison of different genotypic methods for the identification of Achromobacter spp. isolates
| Isolate | Year of isolation | nrdA analysis17 and ST | PCR blaOXA21 | blaOXA19 |
|---|---|---|---|---|
| A39 | 2008 | A. xylosoxidans | + | blaOXA-114m |
| A43 | 2009 | A. dolens | + | blaOXA-787 (MK388909) |
| A45 | 2011 | A. pulmonis | − | – |
| A46 | 2011 | A. marplatensis | − | – |
| A49 | 2012 | A. xylosoxidans | + | blaOXA-790 (MK388911) |
| A50 | 2012 | A. dolens | + | blaOXA-789 (MK388910) |
| A51 | 2012 | A. xylosoxidans | + | blaOXA-114o |
| A52 | 2004 | A. xylosoxidans | + | blaOXA-791 (MK388912) |
| A53 | 2012 | A. dolens | + | blaOXA-364a |
| A79 | 2012 | A. xylosoxidans | + | blaOXA-114i (MK388913) |
| A113 | 2009 | A. xylosoxidans | + | blaOXA-790 (MK388911) |
| A131 | 2012 | A. ruhlandii | + | blaOXA-258a |
| A134 | 2013 | A. ruhlandii | + | blaOXA-258a |
| 38 | 2002 | ST 148 (A. ruhlandii)a | + | blaOXA-258a |
| 39 | 2002 | ST 148 (A. ruhlandii)a | + | blaOXA-258a |
| 67 | 2004 | ST 148 (A. ruhlandii)a | + | blaOXA-258a |
| 79 | 2004 | ST 64 (A. insuavis)a | + | blaOXA-243a |
| 80 | 2004 | ST 14 (A. ruhlandii)a | + | blaOXA-258a |
| 114 | 2002 | ST 165 (A. insuavis)a | + | blaOXA-243e |
| 319 | 1996 | ST 43 (A. ruhlandii)a | + | blaOXA-258b |
| 336 | 2010 | ST 164 (A. dolens)a | + | blaOXA-364a |
| A1 | 2004 | A. xylosoxidans | + | blaOXA-114g |
| A2 | 1995 | A. xylosoxidans | + | blaOXA-114i |
| A3 | 2006 | A. xylosoxidans | + | blaOXA-114f |
| A4 | 2003 | A. xylosoxidans | + | blaOXA-114g |
| A5 | 2002 | A. xylosoxidans | + | blaOXA-114l |
| A6 | 2007 | A. xylosoxidans | + | blaOXA-114f |
| A7 | 2007 | A. xylosoxidans | + | blaOXA-114c |
| A8 | 2008 | A. xylosoxidans | + | blaOXA-114f |
| A9 | 2008 | A. xylosoxidans | + | blaOXA-114l |
| A10 | 2008 | A. xylosoxidans | + | blaOXA-114f |
| A11 | 2008 | A. xylosoxidans | + | blaOXA-114f |
| A12 | 2008 | A. xylosoxidans | + | blaOXA-114e |
| A13 | 2008 | A. xylosoxidans | + | blaOXA-114l |
| A14 | 2001 | A. xylosoxidans | + | blaOXA-114c |
| A15 | 2001 | A. xylosoxidans | + | blaOXA-114c |
| A16 | 2007 | A. xylosoxidans | + | blaOXA-114f |
| A17 | 2006 | A. xylosoxidans | + | blaOXA-114a |
| A18 | 2006 | A. xylosoxidans | + | blaOXA-114a |
| A19 | 2007 | A. xylosoxidans | + | blaOXA-114i |
| A20 | 1996 | A. xylosoxidans | + | blaOXA-114o |
a Isolates identified by ST analysis. Data published previously14,19.
nrdA gene sequencing17 allowed to identify 33/41 isolates. In the remaining isolates, nrdA sequences corresponded to alleles that were not deposited in pubmlst.org/achromobacter. For these isolates, identification was performed based on the complete MLST Scheme18. Different alleles of blaOXA-114 were observed in the 26 isolates identified as A. xylosoxidans by nrdA gene sequencing17; even, 3 new blaOXA-114 variants were identified in this study (accession numbers: MK388911, MK388912, MK388913) (Table 1). These new blaOXA-114 variants were designated as OXA-114i, OXA-790 and OXA-791 by NCBI. Then again, in those isolates identified as A. insuavis, A. dolens and A. ruhlandii by nrdA gene sequencing or ST analysis, the blaOXA sequences displayed 99–100% identity to blaOXA-243, blaOXA-364 and blaOXA-258, respectively. Two new blaOXA-364 variants were identified (MK388909, MK388910) (Table 1). These blaOXA-364 variants were designated as OXA-787 and OXA-789 by NCBI. No blaOXA amplicons could be obtained in any of the isolates identified by nrdA sequencing17 as Achromobacter marplatensis and Achromobacter pulmonis (Table 1).
A. xylosoxidans accounted for 63.4% of Achromobacter colonization/infections in this study, while A. ruhlandii accounted for 17.1%. The remaining species corresponded to A. dolens (9.8%), A. insuavis (4.9%), A. marplatensis (2.4%) and A. pulmonis (2.4%). According to previous studies performed in the UK7, Spain4, France2, USA17, Brazil16 and Denmark11, A. xylosoxidans was the most frequently recovered species from the airways of patients with cystic fibrosis. A. ruhlandii, the second most prevalent species identified in this study, was only identified in 3/96 patients in the UK and none in Spain and France; however it was the second most prevalent species in the USA and Brazil. The reported prevalence for A. dolens varies between 2 to 17%, accounting for 10% in the present study. Discrepant results were observed in species prevalence in different countries, the local distribution being more similar to that reported in Brazil.
All isolates included in this study were resistant to quinolones and aminoglycosides, while piperacillin, piperacillin/tazobactam and carbapenems were the most active antibiotics, as it had been previously described by Almuzara et al.1 Fifty percent (50%) of the isolates were resistant to trimethoprim-sulfamethoxazole while 85% were resistant to colistin. Moreover, 3/7 A. ruhlandii isolates and 1/26 A. xylosoxidans were resistant to imipenem. Antimicrobial MIC values for the Achromobacter isolates included in this study are shown in table S2 (supplementary material).
ConclusionsAlthough A. xylosoxidans was the most common species recovered from the clinical samples of patients with CF, it was not the only species present in those samples, as it was inferred from the phenotypic approaches analyzed in this study. A. ruhlandii was the second species in prevalence, in agreement with the results obtained in Brazil and USA.
MALDI-TOF rendered inconclusive or misleading results understating the presence of species other than A. xylosoxidans, probably due to commercial databases, key components of MALDI-TOF platforms, which are constructed with a low number of isolates mainly corresponding to A. xylosoxidans. Their expansion should be crucial to resolve many of the current inadequate identifications, improving the usefulness of this technique.
On the other hand, concordant results were achieved with nrdA sequencing or ST analysis, and the blaOXA sequencing approach proposed by Traglia et al.19, allowing a reliable discrimination among the different Achromobacter species, and demonstrating that a wide diversity of Achromobacter spp. may colonize and/or infect the airways of patients with CF. Although the MLST scheme constitutes an accurate tool, its arduous procedure may not be applicable in moderate complexity laboratories. In this regard, the amplification and sequencing of a single gene, nrdA or blaOXA, may be more pertinent.
The susceptibility profile of Achromobacter spp. indicated that these microorganisms were resistant to a wide range of antibiotics, including fluoroquinolones, aminoglycosides and the majority of broad-spectrum β-lactams. Carbapenems were the most active antibiotics; however, the emergence of resistant isolates was detected. No correlation could be established among the susceptibility profiles and Achromobacter species. Therefore, the antimicrobial therapy in patients with CF should be conducted based on the susceptibility profile of individual Achromobacter spp. isolates.
Finally, prompt and accurate identification tools should provide an opportunity to understand the clinical impact of the different Achromobacter species on the progression of respiratory infections in patients with CF. In this sense, the development of a robust MALDI-TOF database should be desirable.
Ethical approvalNot required.
Funding sourcesThis work was partially supported by grants from UBACyT to M. Radice and G. Gutkind; ANPCyT to M. Radice, and PIP to G. Gutkind.
M. Radice and G. Gutkind are members of Carrera del Investigador Científico (CONICET). M. Papalia is recipient of a posdoctoral fellowship from CONICET.
Conflict of interestThe authors declare that they have no conflicts of interest.