In the last decade Achromobacter spp. has been associated with chronic colonization in patients with cystic fibrosis (CF). Although Achromobacter xylosoxidans is the most frequent species recovered within this genus, other species such as A. ruhlandii have also been reported in these patients. Descriptions of mobile elements are scarce in Achromobacter and none of them have been originated in A. ruhlandii. The aim of this study was to report the full characterization of a plasmid which was maintained in four clonally related A. ruhlandii isolates. Between 2013 and 2015, nine A. ruhlandii isolates were recovered from a pediatric patient with CF at a hospital in Buenos Aires. Four selected clonally related isolates were sequenced by Illumina MiSeq, annotated using RAST and manually curated. The presence of a unique plasmid of 34096-bp and 50 CDS was observed in the four isolates, displaying only 1 nucleotide substitution translated into one amino acid change among them. These plasmids have a class 1 integron containing the aac-(6′)-Ib gene, a mercury resistance operon region and the relE/stbE toxin/antitoxin system. Plasmids showed 79% similarity and 99% identity with pmatvim-7 from Pseudomonas aeruginosa. This is the first full description and characterization of a plasmid from A. ruhlandii which was maintained over time.
Durante la última década, Achromobacter spp. han sido asociadas con la colonización crónica en pacientes con fibrosis quística. Si bien Achromobacter xylosoxidans es la especie más frecuentemente recuperada, otras especies como Achromobacter ruhlandii también fueron reportadas en nuestra región. Sin embargo, pocos reportes se han centrado en la descripción de elementos móviles, y ninguno de ellos los documenta en A. ruhlandii. El objetivo de este estudio fue reportar la caracterización completa de un plásmido conservado en 4 aislamientos clonalmente relacionados de A. ruhlandii. Se recuperaron 9 aislamientos de A. ruhlandii entre 2013 y 2015 de un único paciente con fibrosis quística proveniente de un hospital pediátrico de Buenos Aires, Argentina. Se realizó la secuenciación completa del genoma de los 4 aislamientos seleccionados según el perfil de resistencia antibiótica en un equipo Illumina MiSeq. Estos fueron anotados mediante RAST y curados manualmente. Se detectó la presencia de un solo plásmido de 34.096pb y 50CDS en los 4 aislamientos, observándose únicamente un cambio nucleotídico traducido en un cambio aminoacídico en un aislamiento. Los plásmidos ensamblados se caracterizaron por presentar un integrón de clase 1 que contenía el gen aac-(6′)-Ib, un operón de resistencia a mercurio y el sistema de toxina-antitoxina relE/stbE. Cabe destacar que estos plásmidos poseen un 79% de similitud y un 99% de identidad con el plásmido pmatvim-7 de Pseudomonas aeruginosa. Esta es la primera descripción y caracterización completa de un plásmido proveniente de A. ruhlandii.
Achromobacter spp. are gram negative rods capable of causing infections, including bacteremia, pneumonia and meningitis, mainly in immunocompromised patients. However, infections in immunocompetent patients have also been reported11,13. In addition, Achromobacter spp. are increasingly recognized as pathogens in patients with cystic fibrosis (CF). Different Achromobacter species, such as Achromobacter xylosoxidans, Achromobacterruhlandii, Achromobacterdolens and Achromobacterinsuavis, have been recovered from these patients, displaying variable frequencies among studies reported in different countries16. Members of this genus are naturally resistant to many antibiotics including ampicillin, cephalotin, aztreonam, cefotaxime, cefoxitin and aminoglycosides. The expression of multiple efflux pumps and the production of chromosomally encoded ß-lactamases1 may be responsible for this resistance profile. Moreover, isolates displaying resistance to clinically relevant antibiotics such as colistin, carbapenem and fluoroquinolones have been described16. Little is known about the resistance markers involved in this acquired resistance profile. Thus, effective antimicrobial therapy can be challenging due to its inherent and acquired multidrug resistance patterns. Only a few plasmid-encoded ß-lactamases have been identified in A. xylosoxidans, as is the case of VEB-115 and class B carbapenemases, such as blaIMP-1, blaIMP-10, blaVIM-11 and blaVIM-21. Moreover, plasmid descriptions are scarce in this genus, and no characterization of plasmids from A. ruhlandii have been reported, possibly due to misidentification of the species belonging to this genus.
Here we report the full characterization of a plasmid which was maintained in four clonally related A. ruhlandii isolates recovered from a single pediatric patient with CF, between 2013 and 2014.
Materials and methodsNine Achromobacter spp. isolates were recovered from a pediatric patient with CF at a hospital in Buenos Aires between 2013 and 2015 (isolation dates are shown in Table 1). The isolates were cultured under aerobic conditions at 37°C on TSA (Tryptic Soy Agar) and accurate identification was achieved by amplification and sequencing of the nrdA gene according to Spilker et al.18XbaI-PFGE was performed according to Mireille-Cheron et al.5 to evaluate the clonal relationship between successive isolates. Minimum inhibitory concentration (MIC) values were determined for a representative set of antibiotics: ampicillin, piperacillin, piperacillin/tazobactam, ceftazidime, cefepime, imipenem, meropenem, ciprofloxacin, levofloxacin, kanamycin, gentamicin and colistin. The antimicrobial susceptibility test was assayed by the agar dilution method according to CLSI recommendations, using the breakpoints established for the category “other non-Enterobacteriaceae”7. The Master pure DNA purification kit (Epicenter, Madison, WI, USA) was used for DNA extraction in four selected isolates (Table 1). These 4 isolates were chosen because, in addition to being clonally related, they displayed differences in their antibiotic susceptibility profiles regarding antimicrobial therapy in patients with CF.
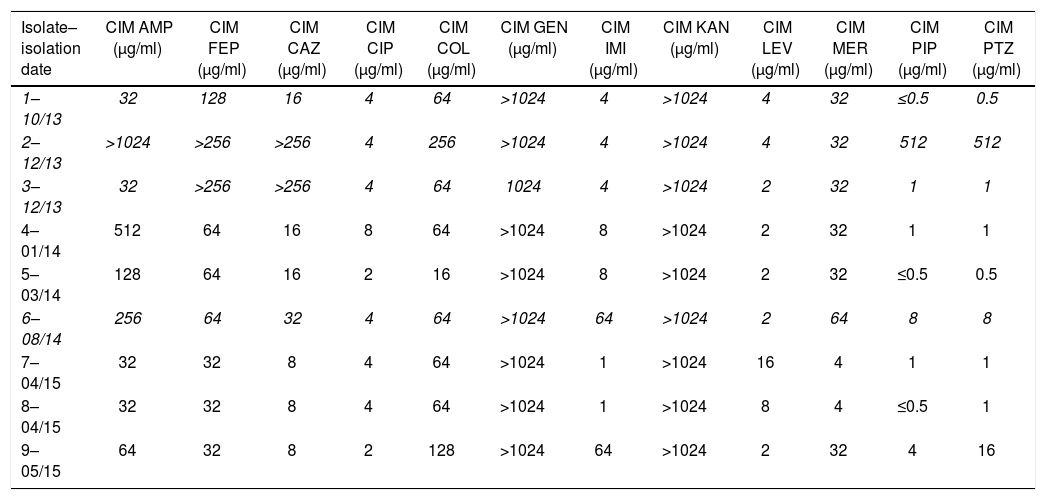
Antimicrobial resistance profiles of the tested isolates.
| Isolate–isolation date | CIM AMP (μg/ml) | CIM FEP (μg/ml) | CIM CAZ (μg/ml) | CIM CIP (μg/ml) | CIM COL (μg/ml) | CIM GEN (μg/ml) | CIM IMI (μg/ml) | CIM KAN (μg/ml) | CIM LEV (μg/ml) | CIM MER (μg/ml) | CIM PIP (μg/ml) | CIM PTZ (μg/ml) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1–10/13 | 32 | 128 | 16 | 4 | 64 | >1024 | 4 | >1024 | 4 | 32 | ≤0.5 | 0.5 |
| 2–12/13 | >1024 | >256 | >256 | 4 | 256 | >1024 | 4 | >1024 | 4 | 32 | 512 | 512 |
| 3–12/13 | 32 | >256 | >256 | 4 | 64 | 1024 | 4 | >1024 | 2 | 32 | 1 | 1 |
| 4–01/14 | 512 | 64 | 16 | 8 | 64 | >1024 | 8 | >1024 | 2 | 32 | 1 | 1 |
| 5–03/14 | 128 | 64 | 16 | 2 | 16 | >1024 | 8 | >1024 | 2 | 32 | ≤0.5 | 0.5 |
| 6–08/14 | 256 | 64 | 32 | 4 | 64 | >1024 | 64 | >1024 | 2 | 64 | 8 | 8 |
| 7–04/15 | 32 | 32 | 8 | 4 | 64 | >1024 | 1 | >1024 | 16 | 4 | 1 | 1 |
| 8–04/15 | 32 | 32 | 8 | 4 | 64 | >1024 | 1 | >1024 | 8 | 4 | ≤0.5 | 1 |
| 9–05/15 | 64 | 32 | 8 | 2 | 128 | >1024 | 64 | >1024 | 2 | 32 | 4 | 16 |
The isolates marked in italic (1, 2, 3 and 6) were selected for WGS.
Libraries were prepared for sequencing using the Illumina NexteraXT kit (Illumina Inc., San Diego, CA), and sequenced using paired-end reads of 150 base pairs in an Illumina NextSeq500 system at the Microbial Genome Sequencing Center. De novo assembly was performed with SPAdes v3.104, using a pre-assembly approach with Velvet v1.2.1021. All possible k-mer lengths were explored. The assembly was annotated using the RAST online server followed by manual curation. The presence of resistance determinants was assessed by Resfinder, MARA and CARD databases10,20.
Plasmids from each isolate were assembled by contig overlapping. The comparison between plasmids was carried out using RAST, BLAST and the Artemis Comparison Tool. One of these plasmids was deposited in GenBank under AN: MK423762.1.
Plasmids from the A. ruhlandii isolates were extracted according to Kado and Liu12. Digestion with KpnI was carried out to compare the restriction profiles. Moreover, PCR amplification of class 1 integron carrying the aac-(6′)-Ib gene was performed6.
To assess their mobilization potential, plasmids were first introduced by electroporation into E. coli DH5α. Transformants were selected in LBA medium containing 32μg/ml kanamycin. Afterwards, conjugation assays were carried out on LBA agar plates using the selected transformants as donor strains and E. coli CAG12177 as recipient cells. Different proportions of donor and recipient cells (10:1, 2:1, 1:1) were spread on agar plates and incubated overnight at 37°C. Transconjugants were selected on agar plates containing 32μg/ml kanamycin and 32μg/ml tetracycline.
ResultsNine clonally related A. ruhlandii isolates were identified from a single patient, based on the results of PCR amplification and sequencing of the nrdA gene, and XbaI-PFGE (data not shown). Different resistance profiles were observed among successive isolates. All isolates were resistant to ampicillin, cefepime, and aminoglycosides, which correlates with the intrinsic resistance profile observed in A. xylosoxidans, and to colistin. Almost all of them were intermediate or resistant to fluoroquinolones. Piperacillin, piperacillin/tazobactam and imipenem were the most active antibiotics. Nevertheless, one A. ruhlandii isolate was resistant to piperacillin and piperacillin/tazobactam and four out of nine A. ruhlandii isolates were categorized as intermediate or resistant to imipenem (Table 1). According to the different susceptibility patterns observed, isolates 1, 2, 3 and 6 were selected for WGS.
All genomes showed a size of approximately 6000000bp with 6000 coding sequences and a GC content of 67.7%. Different antibiotic resistance determinants were found in all 4 genomes corresponding to: 6 putative β-lactamases, 33 putative efflux pump coding genes, 1 aminoglycoside modifying enzyme (aac-(6′)-Ib), the dihydropteroate synthase and chloramphenicol O-acetyltransferase and mutations in gyrA, gyrB, parC, and parD. Among the detected β-lactamase ORFs no differences were observed among the compared isolates. These ORFs corresponded to: (i) blaOXA-258a previously reported as species-specific marker in A. ruhlandii15; (ii) blaaxc, recently described by Fleurbaaij et al.9 in A. xylosoxidans as a novel β-lactamase with carbapenemase activity; and (iii) one molecular class A and three class C non identified β-lactamases. Complete sequences for 2 RND efflux pumps (AxyABM and AxyXY-OprZ) were detected in the 4 genomes and no differences were observed among them. AxyABM displayed 91% identity with that reported in A. xylosoxidans and 97% with that reported in A. ruhlandii17. The complete sequence for axyXY-oprZ coding genes displayed 96% identity with the previously described in A. xylosoxidans2,3. Twelve putative outer membrane coding genes were detected, being identical in all isolates. No Pseudomonas aeruginosa OprD porin homologues were found.
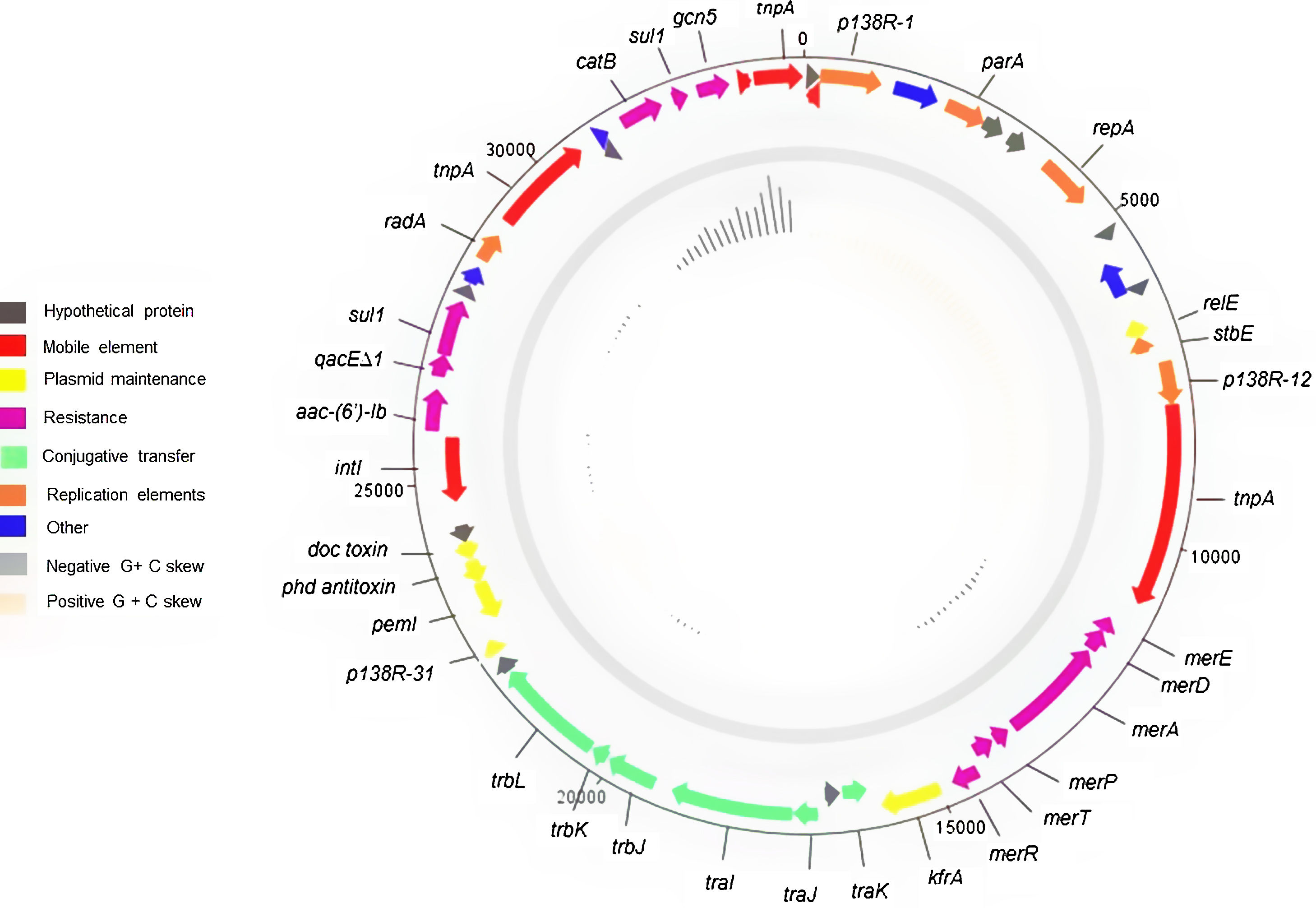
With regard to plasmid characterization, the four identified plasmids had a size of 34096bp with 50 coding sequences and an average G+C content of 63.3% (Supplementary Table 1).
These plasmids contained a mercury resistance operon region of 3351bp containing merT, merR, merP, merA, merD, and merE; which were flanked by a Tn3 family transposase and the tra operon. In addition, it had a partition and replication region of 2217bp comprising genes tnpR, repA, parA and an ATPase gene, which showed 99% coverage and 100% identity with an IncP6 plasmid found in P. aeruginosa (CP033834.1). A transference region of 4923bp containing the trb (trbL, trbK, trbJ) and tra (traI, traJ, traK) loci was observed. These plasmids also contained a class 1 integron of 2835bp carrying the aac-(6′)-Ib gene, the relE/stbE toxin/antitoxin system, two copies of the dihydropteroate synthase gene, -one of them being truncated-, and one copy of the chloramphenicol-O-acetyltransferase (Fig. 1).
The four completely sequenced plasmids were almost identical. Only one of them showed one amino acid difference in both dihydropteroate synthase coding genes. The presence of this plasmid was assumed in the other clonally related isolates, based on the identical KpnI restriction profiles and the presence of the aac-(6′)-Ib gene located in the variable region of a class 1 integron displayed among them.
The overall backbone of the plasmids showed high homology (79% query cover and 99% identity) with P. aeruginosa plasmid pmatVim-7 (AM:778842.1), the tra and replication regions being almost identical as mentioned above (Supplementary Fig. 1).
The conjugation assays rendered negative results under the performed conditions.
DiscussionIn this study we report nine clonally related A. ruhlandii isolates, recovered from the same patient. These isolates displayed different antibiotic resistance profiles, in concordance with previous reports in this species8. Variations observed among these isolates suggest adaptive strategies that could lead to microevolution of resistance patterns in these patients. In silico analysis of A. ruhlandii genomes showed a great diversity of antimicrobial resistance determinants, which reveals the genomic plasticity of Achromobacter spp. and suggests that this genus could be a reservoir of antibiotic resistance markers. However, in the in silico comparative analysis no significant differences were observed among the 4 analyzed genomes, despite their different resistance profiles. Further studies on putative resistance markers identified in the whole genome analysis should be conducted to understand the resistance patterns observed in A. ruhlandii isolates.
This is the first full description and characterization of a plasmid from A. ruhlandii. This plasmid was maintained in nine isolates recovered from the same patient. No significant mutations were found in silico among the four sequenced plasmids, indicating that this plasmid was preserved over time. Only an aminoglycoside resistance marker was found on this plasmid, indicating that mutations and/or variations in chromosomal gene expression should be involved in the resistant patterns observed in the isolates.
A comparative analysis of this plasmid with other 26 Achromobacter spp. available plasmids (A. xylosoxidans, Achromobacter insolitus, Achromobacter denitrificans, Achromobacter pestifer) indicated that the present plasmid from A. ruhlandii was not related with the others. The class 1 integron containing the aac-6′-Ib gene and the mercury resistance operon were detected in 6/26 studied plasmids. Only 2 plasmids corresponding to A. insolitus shared both the replicon and the addition system (relE/stbE), while 2 A. xylosoxidans plasmids harbored only the same replicon. The doc/phD toxin/antitoxin system was not observed in any of the studied plasmids.
Despite its high homology with pmatVim-7 from P. aeruginosa, blaVIM-7 was not present in this plasmid. The metallo-carbapenemase coding gene was carried in a mobile element containing the tnpA15 gene, which may indicate that this element could have been either acquired within P. aeruginosa or lost once the plasmid was incorporated into A. ruhlandii14,19. This description provides new insights into horizontally acquired elements in A. ruhlandii, highlighting its capability of acquiring or transferring genes horizontally from/to other non-fermenters.
The conjugation experiments performed by Li et al. on P. aeruginosa were not successful under laboratory conditions, probably due to the lack of certain genes on the tra and trb regions, such as traF, traG, trbA and trbI14. However, a putative oriT was detected, which could make this plasmid mobilizable. These outcomes are also in accordance with our results, which showed the failure to render transconjugants under the assayed conditions, even if some genes of the tra region such as traR and traG that were missing on the plasmid were encoded within the genome. However, the traG gene was truncated and genes of the trb region such as trbP, trbN, and trbM were missing.
This study provides new insights into this emerging pathogen, which may contribute to develop the necessary knowledge to better understand the persistence in patients colonized/infected by this pathogen and likely contribute to a better management of its evolution.
Data availabilityThe plasmid sequence reported here has been deposited in GenBank under the accession number: MK423762.1.
Conflict of interestThe authors declare that they have no conflicts of interest.
FundingThis research was supported by Universidad de Buenos Aires, Programación Científica UBACyT: 20020150100174BA (2016-2019) to Marcela Radice and by ANPCYT-PICT 1925-2015 to Gabriel Gutkind and by ANPCYT-PICT-2017-3996 to Mariana Papalia.