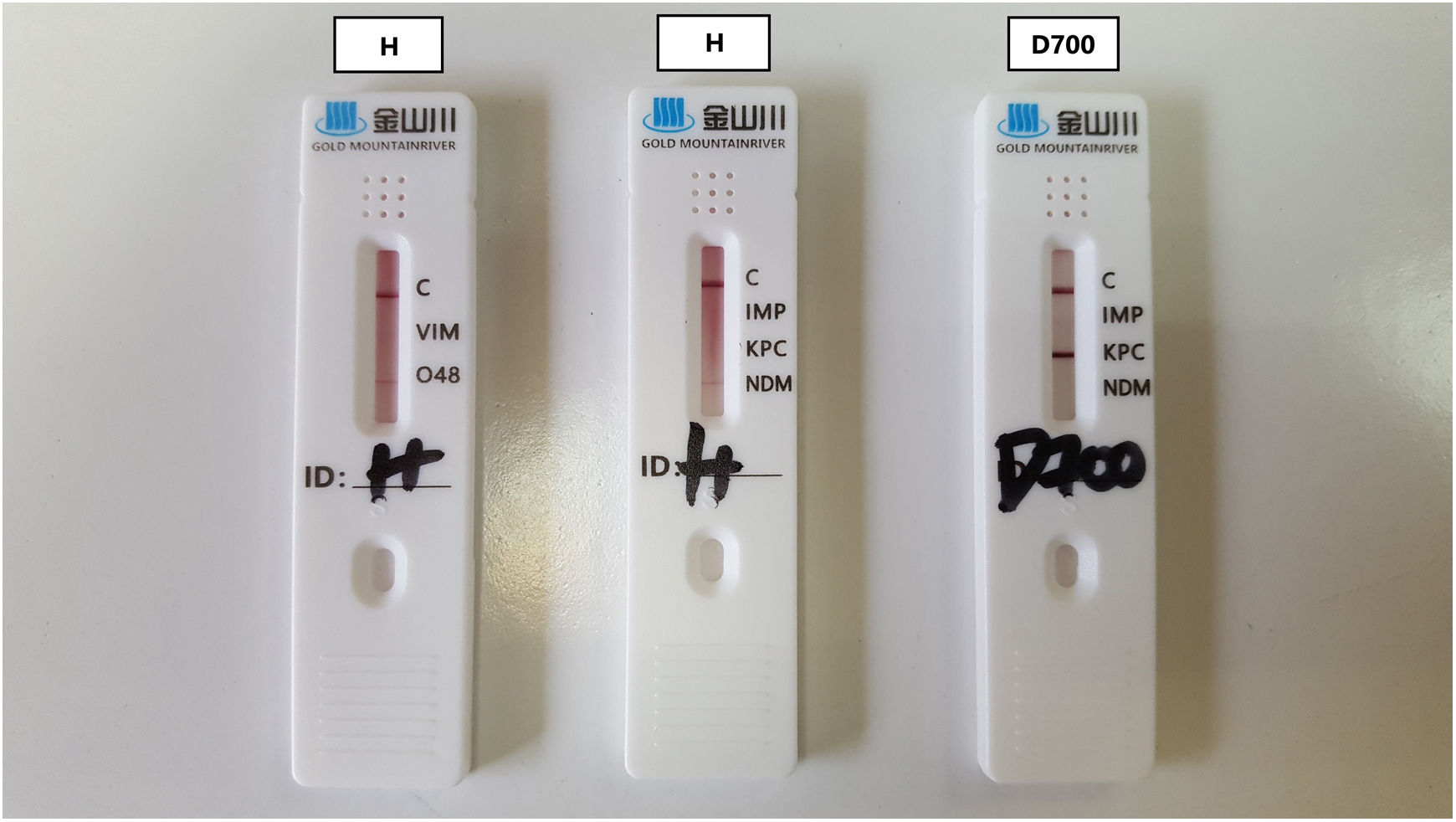
The implementation of lateral flow assays (LFAs) for the phenotypic detection of carbapenemase-production offers faster results than in-house phenotypic methods and lower costs than molecular techniques. However, not enough data are available regarding the performance of each kit, especially among different bacterial species and different carbapenemase targets4.
Surveillance rectal swab cultures for the detection of carbapenem-resistant bacteria are performed in our hospital at patient admission to intensive care units using the Brilliance CRE Medium (Oxoid, Basingstoke Hants, UK). Colonies that grow on that medium are further tested phenotypically for carbapenemase production using the double meropenem disc test5. Whenever a faster result is needed, the carbapenem-resistant K.N.I.V.O. Detection K-set by Goldstream (Beijing, China) is used. This LFA is licensed for the qualitative detection of KPC-, NDM-, IMP-, VIM- and OXA-48 type carbapenemases.
In April 2022, a non-fermenter, Gram negative bacterium with a slow positive oxidase test was isolated from the rectal swab of a female patient. It showed slight bands on the LFA indicating positivity for NDM and OXA-48 carbapenemases (Fig. 1) and was later identified as Stenotrophomonas maltophilia by the VITEK2 automated system (bioMérieux, France). Because of this unusual result, further molecular testing was initially performed using the Novodiag® CarbaR+ kit (MOBIDIAG, Espoo, Finland) that is able to detect blaKPC, blaNDM, blaVIM, blaIMP, various OXA-type and GES-type carbapenemase-encoding genes, mcr-1 and mcr-2. A second molecular confirmation was performed using the semi-automated hybridization system hybriSpot 12™ by Master Diagnóstica (Granada, Spain). This system is able to detect blaKPC, blaNDM, blaIMP, various OXA-type genes, blaGES, blaGIM, blaNMC/IMI, blaSME, blaSIM, blaCMY, blaDHA, blaCTX, blaSHVand blaSIM. Both molecular techniques were negative for all the carbapenemase-encoding genes included in their panel.
S. maltophilia is an opportunistic pathogen of significant concern that can cause nosocomial and community-acquired infections1. Especially for carbapenem and β-lactam resistance, S. maltophilia strains may express among other mechanisms the chromosomally encoded β-lactamases L1 and L2. The presence of blaNDM-1 in S. maltophilia, however, has been sporadically reported in human3 and environmental isolates2.
The positivity of LFAs for the detection of carbapenemases reflects the probable production of the enzyme; therefore, a slight band cannot be ignored as it may indicate the presence of the carbapenemase-encoding gene expressed at lower levels. We believe, though, that this was most probably a false-positive result and the presence of cross-reactivity cannot be excluded. Since it is likely that all LFAs on the market have been verified mainly against Enterobacterales, the users should be very cautious when testing other less frequently encountered bacteria. Even though this was a single case on a pathogen whose intrinsic resistance to carbapenems is already known, it shows the low performance of a rapid detection method in a species which is not usually tested for the presence of carbapenemases due to its intrinsic resistance. Moreover, we tested the LFA with a previously isolated S. maltophilia and a very slight band appeared for OXA-48 but not for NDM. Interestingly, the LFA results for Pseudomonas aeruginosa in our lab are commonly in accordance with the respective phenotypic or molecular tests.
With a very important intervention, EUCAST officially evaluated in the past the performance of all antibiotic-containing discs manufactured by major companies in the industry. A similar official intervention regarding the performance evaluation of different LFAs for carbapenemase detection on various pathogens of clinical significance would also be useful nowadays. For the time being though, we would like to suggest the verification of selected positive LFA results by molecular techniques, especially those showing unusual or novel resistance mechanisms.
Conflict of interestThe authors declare that they have no conflicts of interest.