Our group isolated Salmonella enterica serovar Albany from food and feces of wild captive carnivores in a zoo from northwestern Mexico. This serovar was also associated with the death of an ocelot (Leopardus pardalis) in the same zoo. Another group associated S. Albany with the death of a human patient. It is due to this zoonotic potential that the in vivo study of the host-S. Albany relationship is critical. The recombinant S. Albany-Ovalbumin (rSAO) strain was used to analyze a murine oral infection and its specific cytotoxic T lymphocyte (CTL) response. Our results have shown for the first time that rSAO establishes a systemic infection and evokes epitope-specific lysis with a Th1-like cytokine profile in vivo.
Salmonella enterica serovar Albany fue aislada por nuestro grupo de investigación de alimentos contaminados y de heces de animales carnívoros en cautiverio en un zoológico del noroeste de México; posteriormente, se logró asociar a este serovar con la muerte de un ocelote (Leopardus pardalis), dentro de este mismo zoológico. Otro grupo de investigación asoció a este serovar con la muerte de un paciente. Es debido a este potencial zoonótico que el estudio in vivo de la relación hospedero-S. Albany es crítico. La cepa recombinante S. Albany-Ovoalbúmina (rSAO) fue utilizada para analizar la infección múrida, al igual que la respuesta inmune celular citotóxica específica. Nuestros resultados demuestran, por primera vez, que rSAO establece una infección sistémica y evoca lisis epítopo-específica con un perfil de citocinas tipo Th1 in vivo.
Salmonella enterica serovar Albany (S. Albany) is classified as a non-typhoidal Salmonella. This serovar was previously isolated by our group in food and feces of captive carnivores in a zoo located in northwestern Mexico10. Afterwards, it was also associated with the death of an ocelot (Leoparduspardalis) in the same zoo9. Additionally, this serovar was also involved in a lethal case of a 65-year-old patient11. This zoonotic and lethal potential support our aim to start with the study of important aspects about the host-S. Albany relationship. Here we analyzed the in vivo infection and specific ex vivo CTL immune response using the oral administration of the recombinant S. Albany-Ovalbumin (rSAO) strain in a murine model.
All the reagents used in this study were purchased from Merk (Mexico). Once isolated, S. Albany was transformed with the Ovalbumin (Ova) gene using pST13-OVA, which codifies the OmpC porin from Salmonella Typhi (gently donated by Dr. Ortiz-Navarrete, from CINVESTAV, México) to obtain our recombinant bacterium designated as rSAO. S. Typhimurium 14028 transformed with the same plasmid, designated as rSTO (also donated by Dr. Ortiz-Navarrete), and used as positive control. After the transformation, the Ova expression was confirmed by Western blotting using a homemade mouse polyclonal α-Ova diluted 1:100 as first antibody and rabbit α-mouse IgG conjugated to horse-radish peroxidase diluted 1:3000 as second antibody. The reactions were revealed following protocols previously established in our investigation group using diaminobenzidine and H2O28. Three groups consisting of 3 C57BL/6 mice (6–8 weeks-old) each were formed. While the first group was orally infected using 106 colony-forming units (CFU) of rSAO, the second group was infected with 105CFU of rSTO, due to the fact that S. Typhimurium was lethal for C57BL/6 mice. The mice in the third group were uninfected as previously described2. Six hours post-infection (pi) the feces were collected, weighed, diluted, homogenized in phosphate buffered-saline (PBS) pH 7.2 and plated on brain heart-infusion (BHI) agar supplemented with ampicillin at 100μg/ml; 24h after incubation at 37°C, CFU/mg were determined by colony counting. The mice were monitored every day, weighed, and clinically analyzed according to the scale previously proposed for evaluating mice clinical signs in response to Salmonella infection5. Briefly, the following five clinical criteria were evaluated: general activity level, posture, tremors, fur appearance and finally change in eating habits, which was determined by weighing the food. Each of these aspects was evaluated from a scale of 0–2 for every mouse, where a score of 0 was assigned to severely sick mice, indicating a great presence of the criteria evaluated. This could be evidenced by low activity or a significant decrease in movement, a constant hunch over posture, the presence of tremors, greasy fur appearance and a significant decrease in appetite. Likewise, a score of 1 was assigned to moderately sick mice with a small presence of the clinical sign evaluated. Lastly, a score of 2 corresponded to healthy mice with no change in activity or no signs of sickness5. At day 7 pi, all the clinical criteria values were added. If the final value ranged from 8 to 10, the mice were considered healthy; from 4 to 7, moderately sick; and from 0 to 3, severely sick. Afterwards, the mice were euthanized and their spleens were aseptically removed, measured, and splenocytes were counted using a Neubauer chamber and Trypan Blue 0.4%. To determine systemic infection, splenocytes and bone marrow cells were lysed using 1% TritonX-100, and intracellular bacteria were plated on BHI agar supplemented with ampicillin at 100μg/ml; 24h after incubation at 37°C, CFU were determined by colony counting.
The cytotoxic effector T lymphocytes were obtained by in vitro stimulation with SIINFEKL (Kb restricted OVA-peptide, synthesized by Sigma-Aldrich, Mexico) for 5 days, following standard protocols. Likewise, 1×106 RMA-S cells (also donated by Dr. Ortiz from CINVESTAV-IPN) were stained with CFSE and pulsed with SIINFEKL for 4h prior to the interactions with the cytotoxic effector T lymphocytes (E). RMA-S cells were deemed target cells (T) and effector cells were adjusted to 1:1, 10:1 and 50:1 effector:target (E:T) ratios to evaluate antigen specific lysis; any unspecific lysis was eliminated by normalizing the data with results obtained against an irrelevant peptide (TAVIRADN). The samples were assessed on FACS Accuri C6 (BD) and analyzed using FlowJo software4.
Additionally, the sera of uninfected or infected rSAO or rSTO mice were collected on day 7 pi and were used to determine the cytokine profile associated with the T helper (Th) subset immune response using a CBA Mouse Cytokine Kit (BD). For Th1, the cytokines evaluated were: IFN-γ, TNF-α, IL-2 and IL-6, for Th2: IL-4 and IL-10 and for Th17: IL-17, according to the manufacturer's instructions. The data were acquired on a FACS Accuri C6 (BD) and analyzed using FlowJo software. The data obtained were analyzed using Student's t test on GraphPad Prism 6, where p≤0.05 was considered to be statistically significant.
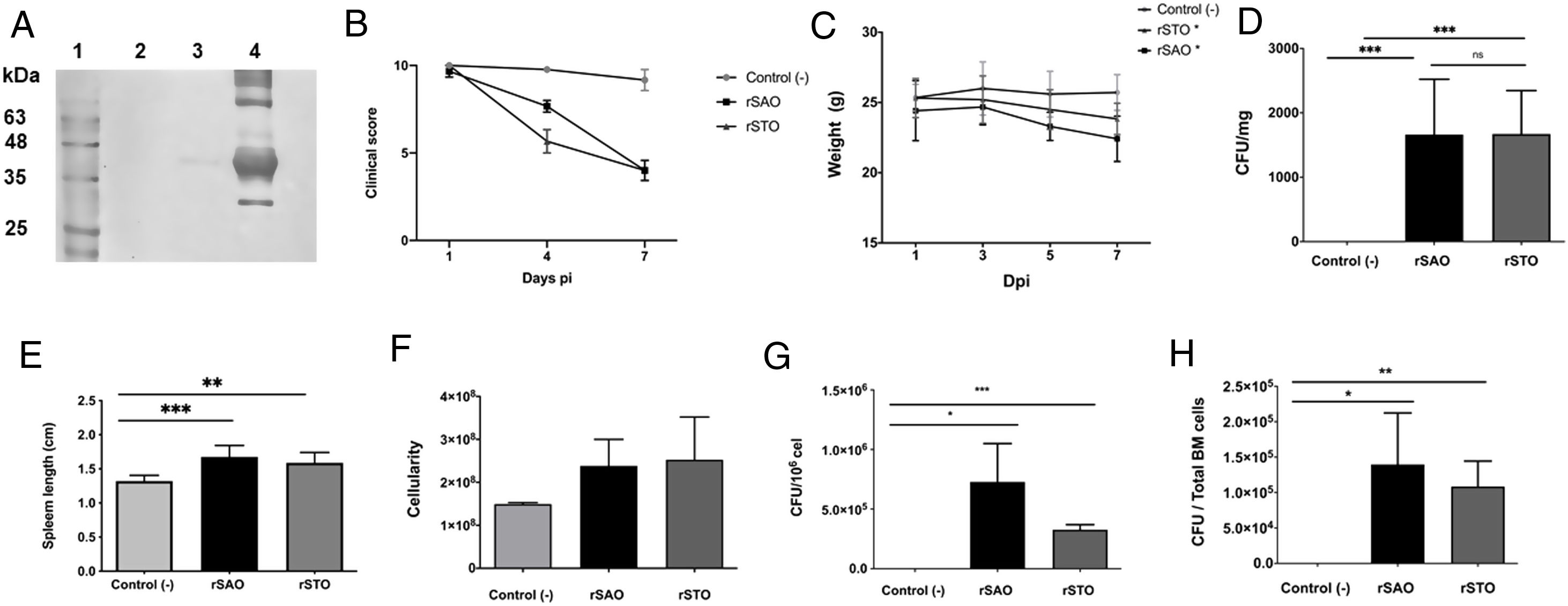
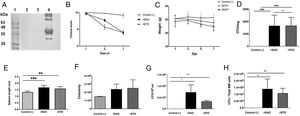
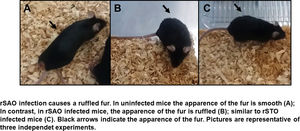
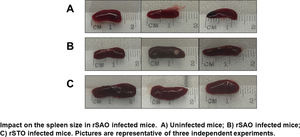
The Ova expression in our recombinant S. Albany strain was confirmed by the Western blot analysis (Fig. 1A), where it was possible to observe a clear band of approximately 45 kDa in crude extract of rSAO, consistent with commercial Ova as positive control in lane 4. The clinical findings of infected as well as uninfected mice are summarized in Figure 1B, and can also be observed in Supplementary Figure 1, where infected mice with rSAO presented a clinical score of 5, which is consistent with that of moderately sick mice, suggesting that this serovar has the capacity to cause characteristic symptoms of an infection by Salmonella. Similar results were obtained from mice infected with the positive control rSTO, while the small differences observed can be explained by the initial inoculum concentration. Both groups presented signs consistent with Salmonella infection, which was considered an initial indicator of the pathogenic potential of rSAO. Various groups have described that along with the behavioral aspects, mice tend to lose weight during infection, and we observed that mice infected with rSAO presented a total average weight loss of 16%, suggesting that the clinical signs observed were significant enough to cause a decrease in appetite in infected mice, and showing that rSAO can display its pathogenic capability (Fig. 1C). To confirm this intestinal infection, the feces collected at 6h pi were plated and the mean CFU observed in rSAO infected mice was of 1650CFU/mg, showing no statistical difference with respect to the rSTO-infected mice (Fig. 1D). These results suggest that rSAO can colonize the intestine, establishing a gastrointestinal infection in mice. Our findings are additionally supported by a similar infection study conducted recently, where it was determined that S. Albany can establish infection and cause pathological changes in tissue in the small intestine, cecum and colon7. To further assess the infection caused by rSAO, the spleens were extracted on day 7 pi and measured; the spleens from mice infected with rSAO presented a mean of 0.6 inches in length, exhibiting a statistically significant difference with respect to the spleens extracted from negative control mice (Fig. 1E). As well as a visible increase in size, it was possible to observe similar macroscopic characteristics in the spleens from mice infected with the positive control rSTO. To demonstrate the possible increased cellularity in infected mice responsible for the increase in spleen size, splenocytes were counted and although the values were not statistically significant, it was evident that there were approximately 0.5 time more cells in the spleens extracted from rSAO and rSTO infected mice when compared to spleens extracted from the non-infected control group (see Figure 1f). Once cellularity was determined, splenocytes were lysed to assess rSAO to evidence its capacity to cause a systemic infection; the mean observed for CFUs in splenocytes of rSAO infected mice was of 3.98×105 per million cells (see Fig. 1G). Parallelly, we determined the bacterial burden in total bone marrow cells to further assess the systemic infection and the mean observed was of 1.87×105 CFUs in total bone marrow cells, demonstrating the capacity of this serovar to establish a systemic infection in mice, thus highlighting its pathogenic potential (see Fig. 1H). A possible explanation, at least in part, for our data, can be related to reports that S. Typhimurium can cause systemic disease in C57BL/6 infected mice, by persisting in B lymphocytes and precursors specifically within the bone marrow1. Maybe rSAO, as well as rSTO, are capable of infecting and establishing a niche in memory B cells and thus, chronic infection. However, more detailed studies should be conducted.
rSAO causes systemic infection in C57BL/6 mice.
(A) Ova expression by transformed S. Albany; Lane 1: molecular weight marker, Lane 2: crude extract S. Albany WT, Lane 3: crude extract rSAO, Lane 4: positive control Ova protein fraction V. (B) Clinical findings in mice orally infected with 1×106 CFU rSAO, 1×105 rSTO and uninfected mice. (C) Weight loss observed in infected and uninfected mice at 1, 3, 5, and 7 days pi. (D) CFU per milligram of feces collected 6h pi. (E) Increase in spleen size measured in cm 7 days pi. (F) Increase in cellularity observed in infected mice spleens 7 days pi. (G) CFU per million cells in infected mice spleens at 7 days pi. (H) CFU in total bone marrow cells at 7d pi. (*) Indicates statistical significance at p≤0.05 using Student's t test. Data are means±SEM (n=3 mice per group) from three independent experiments. rSAO and rSTO are recombinant S. Albany and S. Typhimurium that express the Ovalbumin antigen, respectively.
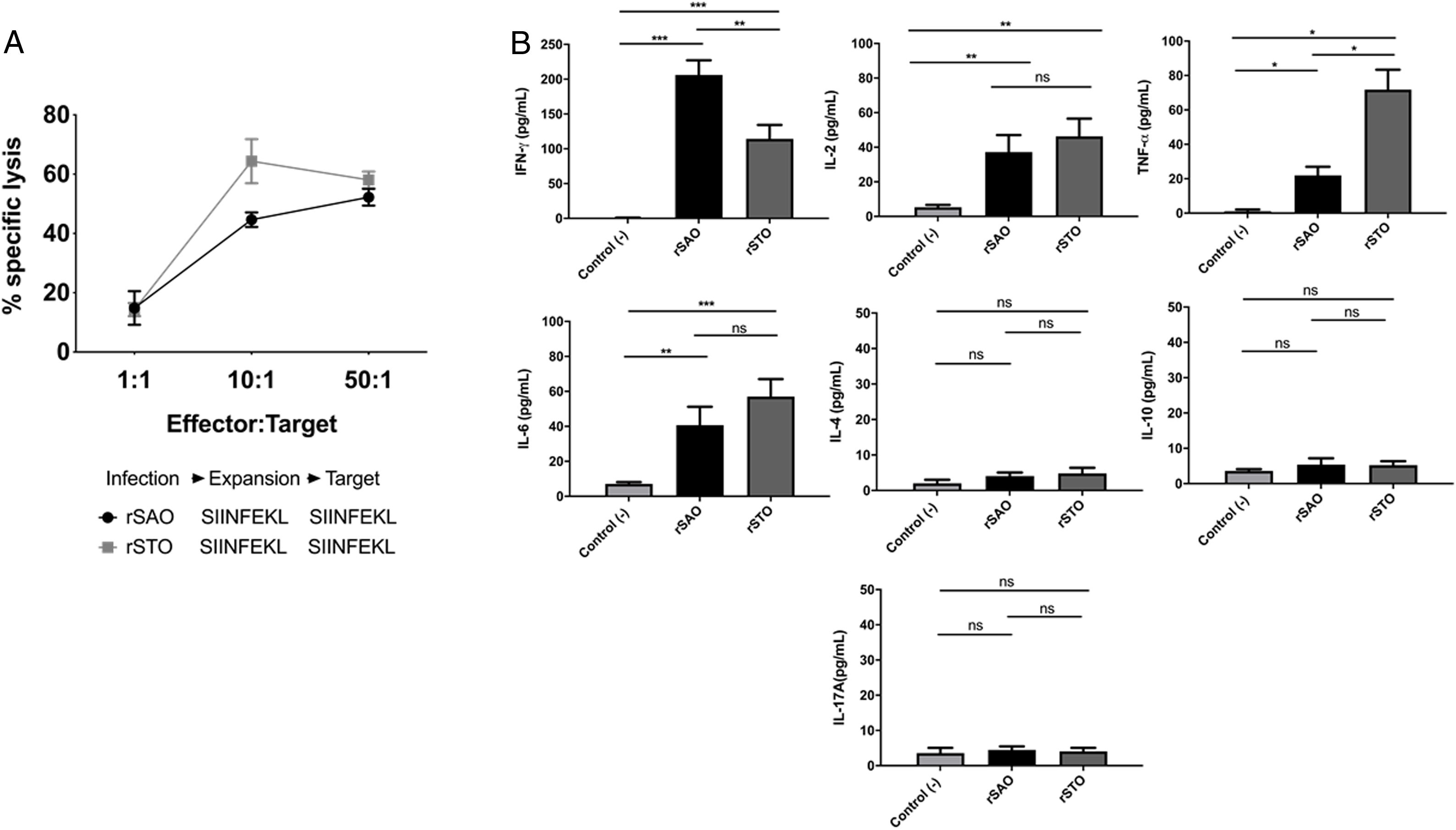
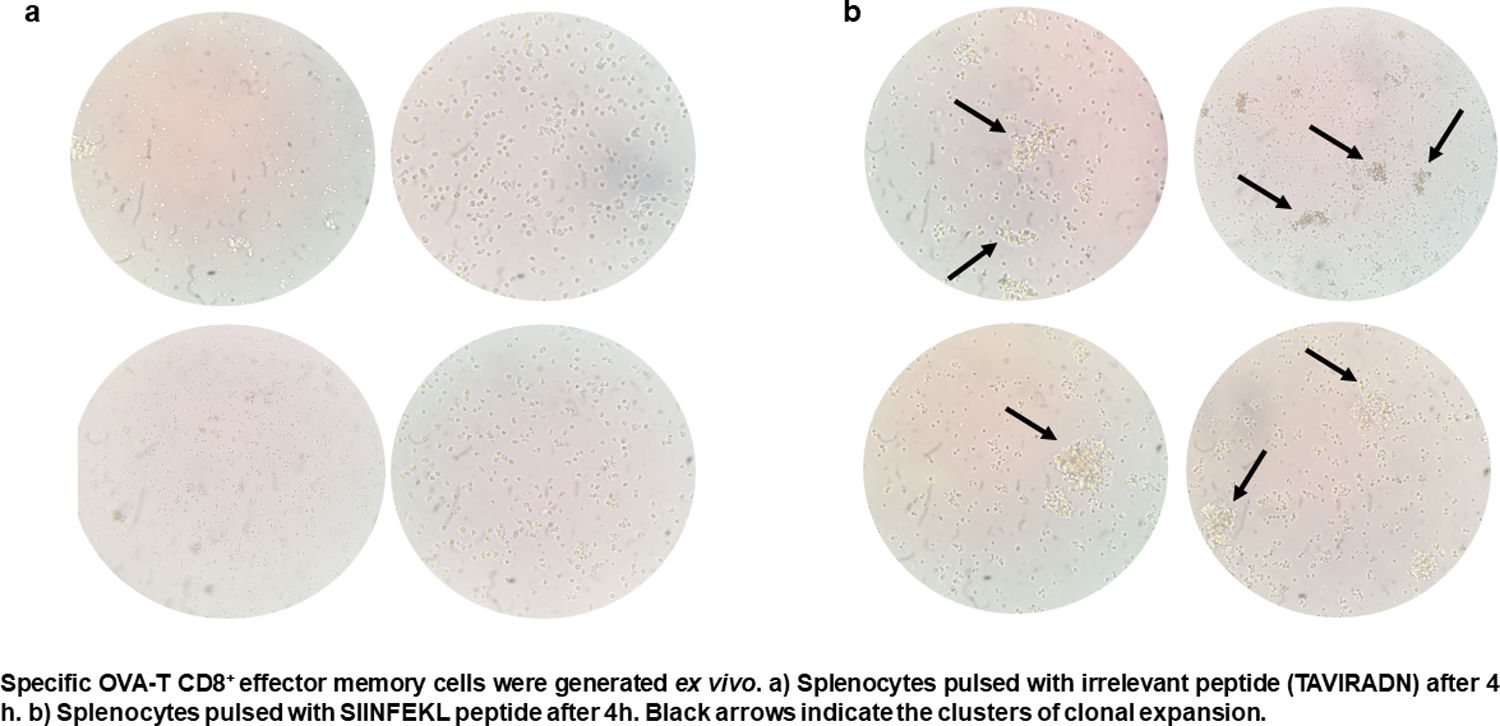
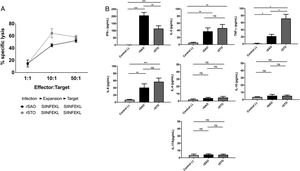
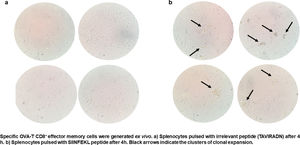
CTL antigen-specific lysis demonstrated that effector cells obtained from rSAO-infected mice were able to kill RMA-S cells pulsed with SIINFEKL, but were inefficient when these target cells were pulsed with an irrelevant peptide. It is relevant to mention that although splenocytes were infected with recombinant bacteria, memory effector T CD8+ clonal expansion was clearly decreased in the irrelevant peptide pulsed group, in contrast with the greater clonal expansion observed in the group pulsed with the SIINFEKL epitope, as shown in Supplementary Figure 3. These results demonstrate that although there is small participation of the genetic construction used, it is not significant. Effector cells evoked 15.6% of specific lysis for 1:1 E:T ratio, 45.1% for 10:1 E:T ratio and 60% for 50:1 E:T ratio. Therefore, CTL from rSAO infected mice recognize this epitope and generate a Kb-restricted CTL response against S. Albany (Fig. 2A). To determine the cytokine profile evoked in vivo by rSAO, the sera of infected and uninfected mice were analyzed, and we observed a predominance of the Th1-like cytokine profile (Fig. 2B). Concentrations of IL-4 and IL-10 were 4 and 5Ypg/ml respectively. IL-6 and IL-2 were both close to 40Ypg/ml, TNF-α concentrations were of 20Ypg/ml, but the IFN-γ concentration was the predominant cytokine, reaching levels of 200Ypg/ml in rSAO-infected mice. These results were consistent with what was observed in our positive control group rSTO, where Th1 cytokines IFN-γ, IL-2, IL-6 and TNF-α were predominant, further suggesting immune response similarities evoked by both serovars. Along with the importance of the Th1 cytokine profile, it has also been reported that the presence of IL-17A along with the Th1 cytokine profile is necessary in controlling Salmonella infection. However, these findings also show that its presence is predominant in the initial stages of infection because its primary function is the recruitment of neutrophils at the initial stages of inflammation2. Because we determined the levels of all the cytokines in the final stages of infection, this could be a possible explanation as to why no statistical difference in the levels of IL-17A was observed across our experimental groups. The secretion of Th1 profile cytokines, specifically IFN-γ, has been previously described in S. Typhimurium infections, where macrophages must be activated to eliminate the intracellular bacteria. Thus, these results suggest that rSAO could establish intracellular infection, evoking protective immunity in mice, which is characterized by the secretion of the Th1 cytokine profile.
rSAO evokes an epitope-specific CTL response with a Th-1 cytokine profile invivo.
(A) Percentages of specific lysis observed at effector:target ratios 1:1, 10:1 and 50:1. Data are means±SEM from three independent experiments. (B) Concentrations of cytokines IFN-γ, TNF-α, IL-2, IL-6, IL-4, IL-10 and IL-17A in sera of infected mice 7 days pi. (*) Indicates statistical significance at p≤0.05 using the Student's t test. Data are means±SEM (n=3 mice per group) from three independent experiments. rSAO and rSTO are recombinant S. Albany and S. Typhimurium that express the Ovalbumin antigen, respectively.
Infection by S. Typhimurium has been thoroughly characterized in C57BL/6 mice and has been used as the comparative gold standard model for other potentially pathogenic Salmonella serovars3. This was consistent with S. Albany, a serovar that had not been previously reported in our region, until its isolation by our investigation group in a zoo from northwestern Mexico, and its further association with the death of an ocelot (Leopardus pardalis) in the same zoo9,10, as it was the first indicator of the pathogenic potential of S. Albany. In this study we have demonstrated for the first time that S. Albany has the pathogenic potential to establish a systemic infection in C57BL/6 mice, displaying the capability to evoke a specific CTL immune response. We found that mice that were orally infected with rSAO exhibited similar nociceptive signs as those infected with the positive control rSTO, suggesting that rSAO has the capability to establish a systemic infection such as rSTO. In addition to these findings, we also demonstrated that rSAO infected mice lost weight in response to the infection, which could be explained by the loss of appetite caused by the strong symptoms induced by the rSAO infection. Likewise, it could also be attributed to the high energy expenditure required to fight off the infection, suggesting that rSAO was capable of establishing a strong enough infection to cause symptoms that directly impacted on the mice appetite and therefore their weight. The capability of rSAO to establish an infection in the gastrointestinal tract was demonstrated by the bacteria isolated from the feces collected 6h pi. The number of colonies exhibited no statistical differences compared with rSTO- infected mice, suggesting that rSAO had the ability to resist stomach acidic conditions and pass through the intestinal mucosa to start infection. This also suggests that rSAO expresses a set of vital virulence factors for establishing infection such as Type III Secretion Systems, Sop and Inv proteins, as well as niche factors that would allow it to overcome bile salts, high osmolarity and competition for nutrients in the intestine; however, to confirm these molecular mechanisms, more detailed studies should be conducted. It has also been previously reported that murine S. Typhimurium infection evokes an increase in spleen size, probably due to the specific clonal expansion of resident immunity cells from this secondary immune organ6.
The specific lysis observed demonstrated that this response was specific for cells harboring the SIINFEKL epitope and CTL were activated by the rSAO infection, generating a specific in vivo immune response. The high concentrations of IFN-γ and TNF-α obtained in our experimental conditions could explain the high percentages of specific lysis as well as the severity of the nociceptive signs observed in infected mice, suggesting that although the infection was strong enough to evoke an immune response, it was probably insufficient to resolve the infection; however, a more detailed study should be conducted to confirm this hypothesis.
We demonstrated here, for the first time that S. Albany causes a systemic disease in C57BL/6 mice and has the same potential to infect other mammalian hosts, including humans, probably expressing a set of virulence factors, pathogenicity islands and secretion systems, although further molecular analyses are needed. Additionally, the isolation of rSAO in bone marrow cells suggest a chronic systemic infection, such as human typhoid fever. The use of recombinant S. Albany-Ovalbumin strain as an antigenic flag represents an important biotechnological tool to study in vivo immune responses. Finally, the CTL immune response evidenced here supports the possibility to manipulate the cellular immune response to eliminate this bacterium effectively.
Authorship contributionCML, PG, SAL, AGE, MAM, EBL, HSLM: Investigation, experimental procedures, data analysis, writing and editing the manuscript. Also, HSLM: design the project, obtained the grant support, and validation of data.
Conflicts of interestNone.
Ethical responsibilitiesNone.
The authors would like to thank fully to Dr. Vianney Francisco Ortiz Navarrete for gently donative the S. Typhimurium 14028 and plasmidic construction pST13-OVA. Also, they extended your acknowledgment to PROFAPI-UAS for financial support with the HSLM's Grant #2015/151. And LBM team for technical assistance