The aim of this study was to evaluate the genotypic relationships among 40 Streptococcus uberis isolated from bovine mastitis by using pulsed-field gel electrophoresis (PFGE). Additionally, the association between PFGE patterns and virulence profiles was investigated. The isolates exhibited 17 PFGE patterns. Different strains were found within and among herds; however, a low number of isolates within the same herd shared an identical PFGE type. No association between PFGE patterns and virulence profiles was found. However, the detection of specific strains in some herds could indicate that some strains are more virulent than others. Further research needs to be undertaken to elucidate new virulence-associated genes that might contribute to the capability of these strains to produce infection.
El objetivo del presente estudio fue evaluar las relaciones genotípicas entre 40 Streptococcus uberis aislados de mastitis bovina mediante la técnica de electroforesis de campos pulsantes (pulsed-field gel electrophoresis [PFGE]). Además, se investigó la asociación entre los patrones de PFGE y los perfiles de virulencia. Los aislamientos mostraron 17 patrones de PFGE. Se encontraron diferentes cepas dentro de los tambos y en los distintos tambos, y un bajo número de aislamientos dentro del mismo tambo compartieron un perfil idéntico de PFGE. No se encontró ninguna asociación entre los patrones de PFGE y los perfiles de virulencia. Sin embargo, la detección de cepas particulares en algunos tambos podría indicar que algunas de ellas son más virulentas que otras. Sería importante avanzar en las investigaciones para identificar nuevos genes relacionados con la virulencia que podrían contribuir a la capacidad infecciosa de estas cepas.
Although an increasing prevalence of Streptococcus uberis mastitis has been reported throughout the world, it has not been conclusively proven that certain clones with enhanced virulence are responsible for mastitis. In an attempt to differentiate S. uberis strains, different typing methods have been developed. Pulsed-Field Gel Electrophoresis (PFGE) is considered a reference technique due to its high discriminatory power and reproducibility. Previous reports have shown evidence of heterogeneity in the S. uberis population using various typing schemes. However, the extent of this is not well defined and its significance to disease pathogenesis has not been determined. Genomic typing of S. uberis was conducted on a number of samples from different countries1–3,5,6,11. Our research group is pioneer in working on molecular typing studies of bovine isolates of S. uberis by PFGE in South America4. As yet, there have been no reports on the association between PFGE patterns and virulence profiles among S. uberis isolates from mastitis in Argentina. The aim of this study was to evaluate the genetic relationships among 40 S. uberis isolates by PFGE and to determine whether certain PFGE patterns are associated with the most frequent virulence profiles. A total of 78 S. uberis isolates were collected from bovine subclinical mastitis from 17 herds located in the central region of Argentina in a previous study7.
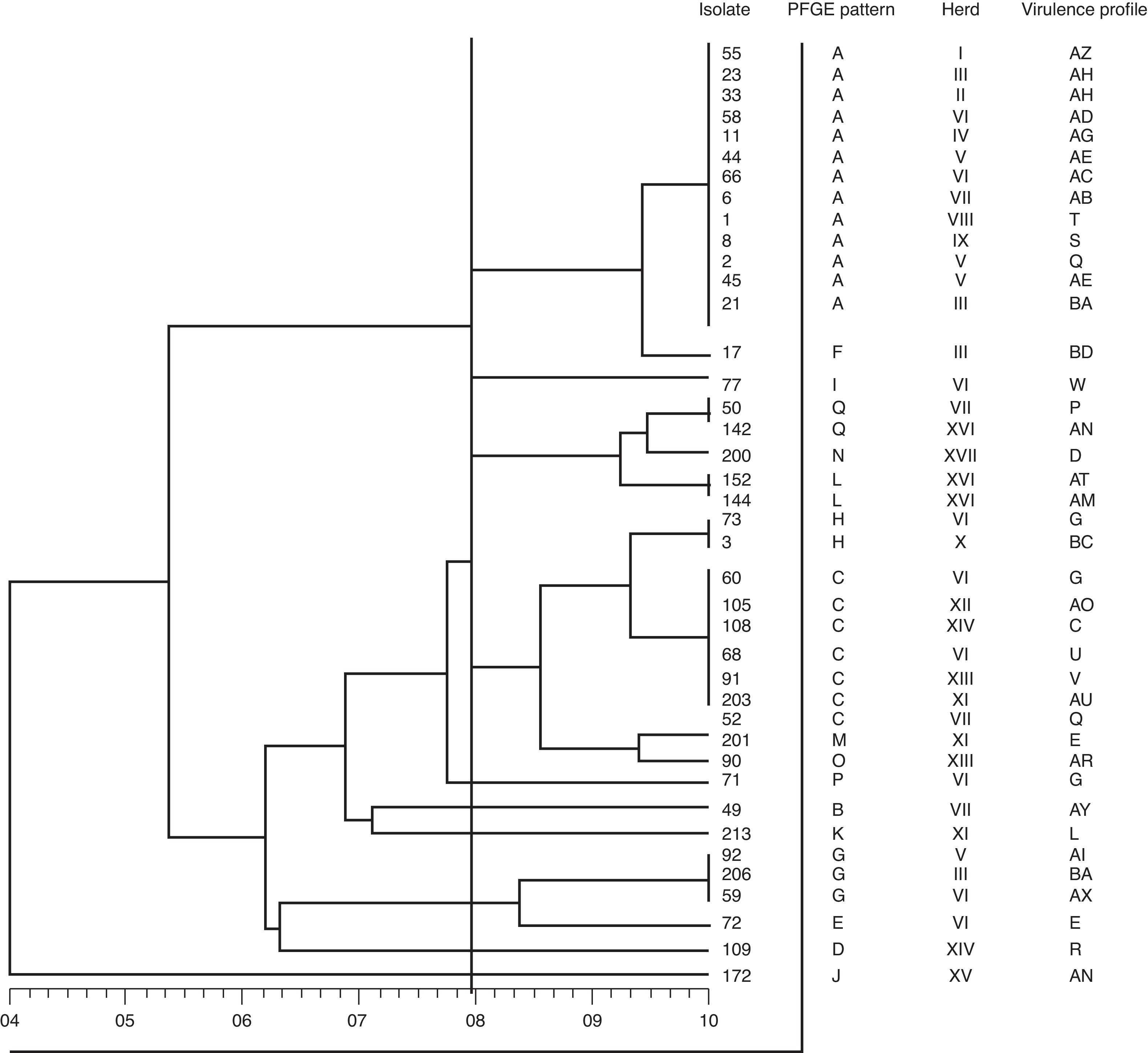
In the present study, forty isolates selected among 78 S. uberis previously characterized7 were used.The distribution of virulence-associated genes as cfu, gapC, hasA, hasB, hasC, lbp, oppF, pauA/B, skc,sua was also determined in the mentioned study. The present work included 15 isolates belonging to the 10 most frequent virulence profiles and 25 isolates belonging to 23 virulence profiles less frequently detected, as previously described7. PFGE typing with SmaI was performed as described by Lasagno et al.4 Briefly, DNA fragments were separated using CHEF-DRII (Bio-Rad) with TBE as running buffer. Electrophoresis was carried out at a voltage gradient of 6V/cm for 20h (14°C) with pulse times increasingly linearly from 1 to 20s. Lambda ladder PFGE markers (50–1000kb; Promega) were included as molecular size standards. On completion of the assay, the gel was stained with 0.5μg/ml of ethidium bromide and banding patterns were visualized using the MiniBisPRO gel documentation system (Micro Photonics Inc., Allentown, USA). Patterns obtained were subjected to the guidelines for interpretation of PFGE based on differences in banding patterns posed by Tenover et al.9 A cutoff value of 80% of genetic similarity among isolates was chosen for discrimination between distinct clusters. Dendrograms were generated by using the Dice coefficients and clustering was done by the unweighted-pair group method with arithmetic averages (UPGMA). Associations between PFGE patterns and virulence genes were examined by the chi-square test. The obtained PFGE patterns yielded 7–10 distinctive fragments from 23 to 340kb size range. The assays were repeated twice. Among the 40 isolates, 17 different PFGE patterns (A–Q) were observed. DNA restriction with SmaI displayed both identical and non-identical PFGE patterns. To determine the degree of similarity of the 17 different PFGE patterns, the 40 isolates were examined by dendrogram analysis. All the isolates were divided into 10 major groups, using a cutoff value of 80% (Fig. 1). Pattern A was the most common, comprising 13 isolates (32.5%) distributed among 9 herds. A similar result was observed for pattern C, which clustered seven isolates (17.5%) distributed among 6 herds. The remaining 15 patterns were less frequent and clustered from 1 to 3 isolates. No evidence of an identical PFGE pattern in different herds was reported by most of other PFGE-based S. uberis epidemiological studies1,5,10,11. Table 1 shows the PFGE pattern distribution of the 40 S. uberis isolates on each dairy farm. Different strains were found within and among herds. Of the 17 herds only herd VI had nine isolates, which were clustered in 7 PFGE patterns. Pulsotypes A and C comprised 2 and 2 isolates, respectively (Table 1). Within herds VII, XI, XII, XIII and XIV no common PGFE patterns were found. The results obtained in this study show that S. uberis is a highly heterogeneous species with multiple PFGE patterns present in a single dairy herd, suggesting that the species is behaving as an opportunistic pathogen and might cause mammary gland infection due to the environmental contamination of the gland. Similar results were demonstrated in previous studies1–3,5,6,11 conducted on cow milk samples from dairy herds located in different regions. Identical results have been recently reported for the first time in Argentina by our research group4. In the present study there were three isolates of identical PFGE types (pulsotype A) recovered from herd V, suggesting that this clone is predominant in this herd. Although the number of isolates of S. uberis examined in each herd was too small in some cases to draw firm conclusions, these data suggest the presence of a single clone that was transmitted either between cows or acquired from a common source. They support the findings from previous studies1,5,10,11, which reported that these events seemed to be uncommon. Phuektes et al.6 also reported the occurrence of a genetically diverse population; however, the authors demonstrated that two strains predominated at higher prevalence (>20%) in two of the four herds. A study carried out by Shome et al.8 was aimed to investigate the virulence characteristics of streptococci from bovine milk and to assess the molecular epidemiology and population structure of the Indian isolates using multilocus sequence typing (MLST) and PFGE. The comparative analysis of MLST and PFGE results indicated that isolates belonging to the same sequence type exhibited similar or related PFGE patterns for both S. agalactiae and S. uberis although restricted to a specific herd. From our results, it can be inferred that specific strains belonging to PFGE patterns A and C in some herds could have some advantages of infecting likely due to a special combination of virulence factors. In a previous report we examined 11 virulence-associated genes by PCR7. We found 58 different virulence profiles, and 33 (42.3%) S. uberis isolates belonged to the 12 most frequent profiles. The diversity of virulent profiles of selected isolates does not allow to draw firm conclusions on the diversity of virulent genes detected among the PFGE patterns. However, the data show the presence of hasC, sua, cfu, and gapC genes in most of the strains belonging to patterns A and C. Our data reveal that most of the isolates (62.5%) had different virulence profiles. In this sense, A and C PFGE patterns grouped 50% of the isolates and showed 11 and 7 different virulence profiles, respectively. A possible relationship between virulence profiles and PFGE patterns was examined by the chi-square test and no association was found (p>0.05). In a previous study, Wang et al.11 did not report the possible existence of clones with enhanced virulence; however, these authors speculated that the great genetic diversity of S. uberis may indicate that virulence is not associated with any specific molecular type. Considering that no association was found between the virulence profiles and PFGE patterns in this study, four isolates were the exception. S. uberis strains 44 and 45 isolated from herd V and designated as the same strain by PFGE (pattern A) had a common virulence profile (AE). Additionally, only two strains (33 and 23) isolated from different herds (II and III) sharing the same PFGE pattern (A) were found to have the same virulence profile (AH). The present study describes the S. uberis genotypes responsible for the mastitis cases and shows that no association between virulence profiles and PFGE patterns was found among S. uberis isolates recovered from subclinical mastitis. Although there is no predominant PFGE pattern, the existence of specific strains in some herds belonging to more frequent patterns, could indicate that some strains may be more virulent than others. Further research needs to be undertaken to elucidate new virulence-associated genes that might contribute to the capability of these strains to produce infection.
The authors declare that no experiments were performed on humans or animals for this study.
Confidentiality of dataThe authors declare that no patient data appear in this article.
Right to privacy and informed consentThe authors declare that no patient data appear in this article.
Conflicts of interestThe authors declare that they have no conflicts of interest.