The fall armyworm, Spodoptera frugiperda (J.E. Smith) (Lepidoptera: Noctuidae), is an important maize pest. Due to the environmental impact and emergence of resistance caused by chemical pesticides and transgenic events, the use of baculoviruses becomes a safe and useful alternative for its control in integrated pest management strategies. Here we report the identification of a novel isolate of a granulovirus of S. frugiperda native to the central region of Argentina, named SfGV ARG. We observed that larvae infected with SfGV ARG showed a yellowish coloration, swollen body and, in some cases, severe lesions in the last abdominal segments. We confirmed the identity of the isolate by sequencing fragments of the lef-8, lef-9 and granulin genes and by calculating evolutionary distances using the Kimura-2-Parameter model. SfGV ARG DNA restriction pattern allowed to estimate a genome of at least 135 kb.
La oruga militar tardía, Spodoptera frugiperda (J.E. Smith) (Lepidoptera: Noctuidae), es una plaga importante del maíz. Debido al impacto ambiental y a la aparición de resistencia causados por los pesticidas químicos y los eventos transgénicos, el uso de baculovirus resulta una alternativa útil y saludable para su control en estrategias de manejo integrado de plagas. En este trabajo reportamos la identificación de un nuevo aislamiento del granulovirus de la S. frugiperda nativo de la región central de Argentina, SfGV ARG. Se observó que larvas infectadas con SfGV ARG mostraron coloración amarillenta, hinchazón y, en algunos casos, lesiones graves en los últimos segmentos abdominales. Se confirmó la identidad del aislamiento por secuenciación de fragmentos de los genes lef-8, lef-9 y granulina, y por cálculo de distancias evolutivas usando el parámetro de Kimura-2. El patrón de restricción generado con el ADN genómico de SfGV ARG permitió estimar un tamaño de genoma de al menos 135kb.
The fall armyworm Spodoptera frugiperda (J. E. Smith) (Lepidoptera: Noctuidae) is a major maize crop pest. The control of this pest is achieved by the use of chemical pesticides and transgenic crops expressing Bacillus thuringiensis (Bt) insecticidal genes. However, due to the emergence of resistance caused by transgenic events, additional synthetic insecticide applications are needed to control S. frugiperda1. Therefore, the use of new control measures becomes an important issue. In this context, baculoviruses are one of the alternatives to control specific insect pests, with a very low risk of emergence of resistance in the field7. Moreover, baculoviruses are highly specific insect viruses with great persistence in the environment due to the occlusion bodies (OBs) that enclose and protect the virions2.
Baculoviridae members are rod-shaped viruses with double-stranded large circular DNA genomes of 80–180 kb9. Initially, this family was classified into two genera, according to the morphotype of their OBs: nucleopolyhedrovirus (NPV) and granulovirus (GV). At present, baculoviruses are classified into four genera: Alphabaculovirus and Betabaculovirus are lepidopteran-specific NPV and GV, respectively; Gammabaculovirus are NPV that infect hymenopterans and Deltabaculovirus comprising dipteran-specific NPV9.
In this study a granulovirus (Betabaculovirus) was isolated from S. frugiperda in an alfalfa crop in the central region of Argentina, which was designated as SfGV ARG. In order to characterize this virus and confirm its identity, three-day-old S. frugiperda larvae were orally infected to amplify the virus and get evidence of infection symptoms. Infected larvae died after at least ten days regardless of the dose used, and on occasions death occurred after about 20 to 30 days, suggesting that SfGV ARG is a type 1 granulovirus due to the slow killing time in comparison to type 2 GV. Infection symptoms included a yellowish-orange coloration and a swollen larva body (Fig. 1). Typically, dead larvae did not show liquefaction, suggesting the absence of the cathepsin gene, as it happens in the Colombian isolate SfGV VG0084. Furthermore, in some cases dead larvae showed conspicuous severe injuries in the last abdominal segments (Fig. 1a, b, e). Samples of these injuries were observed using dark field microscopy where plenty of granulovirus OBs were apparent (not shown). Betabaculoviruses are classified as type 1, 2 or 3: type 1 are slow killing viruses, generally infecting the fat body of the larva whereas type 2 are polyorganotropic, fast killing viruses5. Available data regarding other SfGV isolates suggest that this is a type 1 betabaculovirus: Colombian and Brazilian SfGV isolates have been studied showing mean times to death (MTD) of 29 days and 33 days, respectively, confirming that this virus is a slow- killing betabaculovirus3.
Symptoms shown by S. frugiperda larvae infected with SfGV ARG at different times post infection and with different doses: 17 dpi, 1 × 104 OB/ml (A); 12 dpi, 1 × 106 OB/ml (B); 21 dpi, 1 × 104 OB/ml (C); 14 dpi, 1 × 108 OB/ml (D); 14 dpi, 1 × 108 OB/ml (E); 28 dpi, 1 × 109 OB/ml (F); 14 dpi, uninfected control. dpi: days post infection. Black bar: 0.5cm. Red arrows indicate injuries in the last abdominal segments.
Typical signs of larvae infected with type 1 granuloviruses include markedly swollen larva body and a creamy yellow appearance, as a consequence of infected fat body proliferation5. An early report of S. frugiperda infected by a GV described the diseased larvae as puffy, with a lighter color than normal and with an enlarged fat body, which was much whiter than normal after dissection8. Our observations were in agreement with these signs; however, we also noted (Fig. 1) that this whitish fluid was released from infected larvae in the late stages through an important injury that is naturally produced in the last segments of the abdomen. This observation is different from the typical liquefaction signs described for larvae infected with baculoviruses containing the cathepsin gene in their genomes, which leads to the complete disintegration of the infected larval body10. However, the release of OBs from the dead bodies of SfGV-infected larvae could be enabled by the cuticle rupture produced by the extensive proliferation of the fat body, thereby facilitating the dispersal of the OBs for the horizontal transmission of the virus.
Subsequently, SfGV ARG was replicated by oral infection of S. frugiperda larvae and the produced OBs were purified and subjected to SDS PAGE. The SfGV protein profile shows a typical 30kDa-band that is coincident with the molecular weight of the major OB protein, granulin (29kDa) (Supplementary file 1).
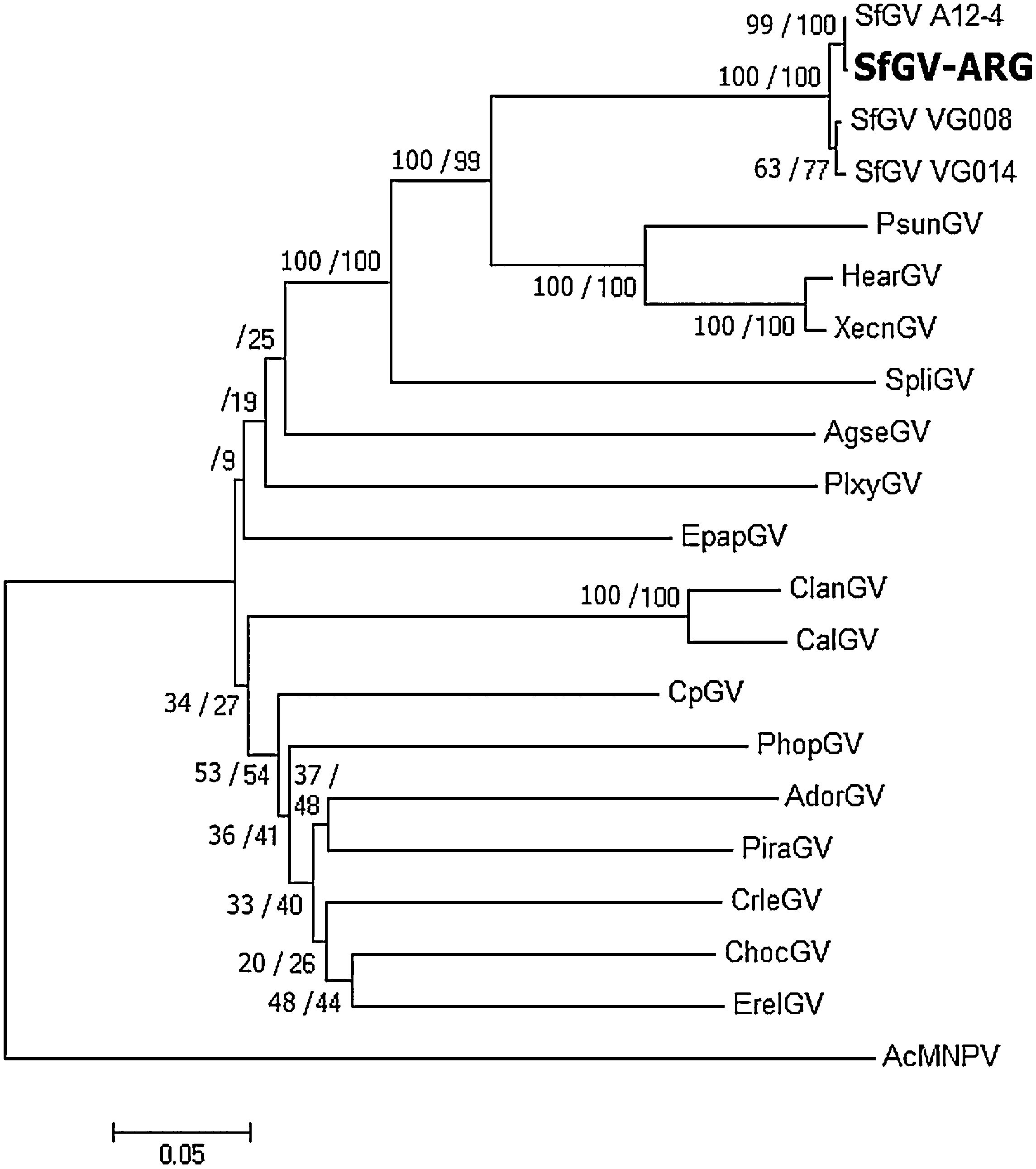
In order to recognize the identity of this isolate, DNA was extracted from purified OBs isolated from dead larvae and subjected to PCR by using degenerate primers that detect the conserved baculovirus genes lef-8, lef-9 and polh/gran as previously described6,11. Blastn searches of these sequences had best hits with other SfGV isolates. Multiple alignments of concatenated partial sequences of the selected genes with those of other betabaculoviruses (Supplementary file 2) were used to infer phylogeny. The phylogenetic tree depicted in Fig. 2 shows that SfGV ARG clusters together with the other 3 SfGV isolates, being SfGV A12-4 (found in a French collection)11 evolutionarily the closest to SfGV ARG.
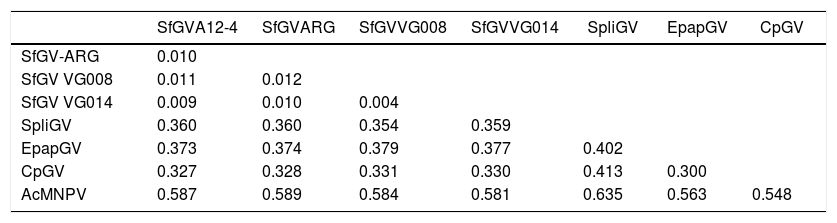
Jehle et al.11, proposed that two (or more) isolates belong to the same “baculovirus species” if the Kimura-2-Parameter distance between single and/or concatenated polh/gran, lef-8 and lef-9 nucleotide sequences is smaller than 0.015. Furthermore, two viruses should be considered a different virus species if the distance between single and/or concatenated sequences is larger than 0.050. For distances between 0.015 and 0.050, complementary information is needed to determine whether two viruses are the same or different species11. K-2-P values obtained for the four SfGV isolates are below 0.015, indicating that they belong to the same species. Table 1 also shows that K-2-P values above 0.050 for SfGV compared with SpliGV, CpGV, EpapGV and AcMNPV, which were chosen as controls for species demarcation.
Evolutionary distances determined with concatenated polh/gran, lef-8 and lef-9 nucleotide sequences using Kimura-2-Parameter
| SfGVA12-4 | SfGVARG | SfGVVG008 | SfGVVG014 | SpliGV | EpapGV | CpGV | |
|---|---|---|---|---|---|---|---|
| SfGV-ARG | 0.010 | ||||||
| SfGV VG008 | 0.011 | 0.012 | |||||
| SfGV VG014 | 0.009 | 0.010 | 0.004 | ||||
| SpliGV | 0.360 | 0.360 | 0.354 | 0.359 | |||
| EpapGV | 0.373 | 0.374 | 0.379 | 0.377 | 0.402 | ||
| CpGV | 0.327 | 0.328 | 0.331 | 0.330 | 0.413 | 0.300 | |
| AcMNPV | 0.587 | 0.589 | 0.584 | 0.581 | 0.635 | 0.563 | 0.548 |
In order to further study the SfGV ARG genome, digestion patterns with EcoRI, HindIII and BamHI were carried out. Banding patterns shown in Supplementary file 3 allowed us to further estimate the size of this large dsDNA genome, which was typical for baculoviruses. Taking into account the EcoRI restriction profile, which shows the largest number of bands with least overlapping compared to the BamHI and HindIII profiles, we estimated a genome of at least 135kb by adding up the restriction fragment sizes, which is smaller than the Colombian isolate (140kb)4. However, due to the poor resolution of small bands, it is possible that the restriction fragments under 2000bp were missed in this assay and therefore the genome size might be underestimated.
Although SfGV is not a good candidate to be used alone as a biocontrol agent, it has proved to be a potential enhancer of NPV infections when used in viral mixtures12, probably because it encodes two enhancin proteins in its genome, as in the case of the Colombian isolate. These features remain to be demonstrated for SfGV ARG. The study of different baculovirus isolates is of great interest for the application of biocontrol strategies against the fall armyworm in the Americas and also in African countries where its presence has been recently reported13.
FundingThis work was supported by grants from the Agencia Nacional de Promoción Científica y Tecnológica (ANPCyT, PICT 2013-0762).
Conflict of interestThe authors declare that they have no conflicts of interest.
The authors thank Alicia Sciocco-Cap for assistance with SfGV isolate and for thoroughly revising the manuscript. Also the authors thank Silvana Tongiani and Paula Giménez for the maintenance of the insect colony at IBBM.