The possibility to obtain DNA from smears is a valuable alternative to remedy the lack of samples when they are totally used for bacilloscopy; this technique solves the biosafety problem related to a possible accident with the transportation of flasks containing potentially transmissible clinical samples. Hence, the purpose of this study was to utilize the insertion sequence IS6110 for amplification of DNA from a smear-positive sample for tuberculosis (TB) diagnosis. Among the 52 positive bacilloscopies, sensitivity, specificity, positive predictive value and negative predictive value were 52.3%, 100%, 100% and 89.7%, respectively whereas accuracy was 90.7%. The IS6110-based PCR for TB diagnosis developed in DNA extracted from a positive smear is a fast, simple, specific, and safe method.
La posibilidad de obtener ADN a partir de frotis es una valiosa alternativa para remediar la falta de muestras cuando estas son totalmente utilizadas para la baciloscopia; esta opción soluciona, además, el problema de bioseguridad asociado a la posibilidad de accidente al transportar frascos que contienen muestras clínicas potencialmente infectivas. Por lo tanto, el propósito de este estudio fue utilizar para el diagnóstico de la tuberculosis la secuencia de inserción IS6110 para amplificación del ADN a partir de frotis que resultaron positivos por baciloscopia. Del análisis de 52 baciloscopias positivas surge que la sensibilidad, la especificidad, el valor predictivo positivo y el valor predictivo negativo de esta técnica fueron, respectivamente, del 52,3%, del 100%, del 100% y del 89,7%; y la precisión fue del 90,7%. La PCR IS6110 para el diagnóstico de tuberculosis, desarrollada con ADN extraído de frotis positivos, es un método rápido, simple, específico y seguro.
Tuberculosis (TB) continues to be a large global health problem. In 2011, there were 84,137 notified TB cases in Brazil, 40,289 of which showed positive-bacilloscopy, accounting for 66% of pulmonary TB; 12,683 new cases presented negative bacilloscopy findings, and 10,067 were extrapulmonary TB cases. In view of such data, Brazil currently ranks 15th on the list of 22 countries presenting the highest TB indexes all over the world12. Nontuberculous Mycobacteria (NTM) also have the capacity to cause the disease, and their importance is progressively growing with the isolation of different mycobacterial species in the laboratory. The clinical-laboratory correlation is of crucial importance for the diagnosis and determination of a therapeutic strategy4. Bacilloscopy is a simple, fast, and inexpensive test, being the most commonly used technique for TB diagnosis. The possibility to obtain DNA from smears is a valuable alternative to remedy the lack of samples when these are totally utilized for bacilloscopy; in addition, this technique solves the biosafety problem related to the transportation of flasks with clinical samples that are potentially transmissible, in case of an accident2,9,10. The amplification of nucleic acids by means of PCR directly from smears has been described by some authors as more specific with respect to bacilloscopy and faster than culture methods2,10. Positive bacilloscopy does not differentiate Mycobacterium tuberculosis complex (MTBC) species from Nontuberculous Mycobacteria (NTM) ones; however, when associated to PCR, it increases the accuracy of TB diagnosis5,8. The goal of the present study is to use the insertion sequence IS6110 for amplification of DNA from smears for TB diagnosis and to exclude it when the smear is positive, taking biosafety into account.
Two hundred eighty-seven smears (convenience samples) coming from the Faculty of Medicine/Laboratory of Research of Mycobacteria/University Hospital – Federal University of Minas Gerais (Faculdade de Medicina/Laboratório de Pesquisa em Micobactérias/Hospital das Clínicas Universidade Federal de Minas Gerai – FM/LM/HC-UFMG) were selected and stained by the Ziehl Neelsen and Auramine method, along the period between 2010 January and 2011 December. Only those smears having culture results were included. Of the 287 smears, 26 were excluded because they did not meet the inclusion criteria; therefore 261 smears remained. Fifty-two (52) of the smears presented positive bacilloscopies and cultures whereas 35 had negative bacilloscopies and positive cultures and 174 negative bacilloscopies and cultures. Seventy-two (72) of them were identified as MTBC and 15 as NTM. The culture was developed in Löwenstein–Jensen medium and the identification test used phenotypic methods6. DNA extraction was done by Chelex-100+Nonidet P40 (NP40) in smear where a volume of 25μl of Chelex solution, containing 5% Chelex-100, 1% Nonidet P40, 1% Tween 20, and distilled water was added on the smear. The smear was scraped off with the help of a tip, up to complete removal; then, it was transferred to an Eppendorf tube containing 75μl of the same solution. After stirring, the Eppendorf tube was incubated for 30min at 100°C. The samples were centrifuged for 10min at 13,000×g; then, the supernatant was transferred to a new sterile Eppendorf tube and 5μl of the solution was utilized for PCR1,10.
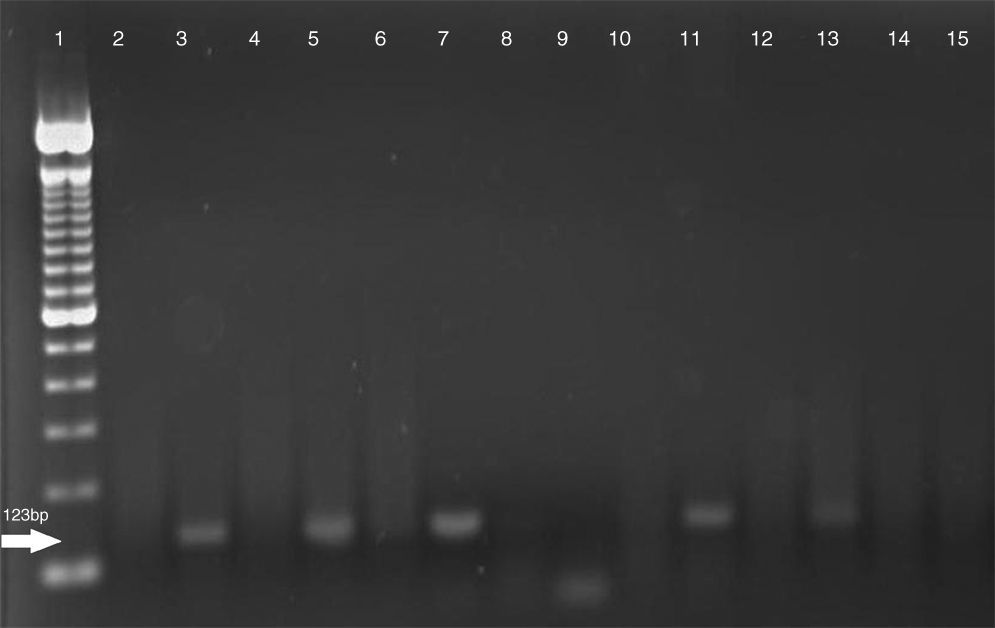
The PCR was developed under the following conditions: 7.0μl of buffer (10×), 3.0μl of 50mM MgCl2, 0.2μl 25mM DNTP, 10pmol of every oligonucleotide (IS1-5′ CCT GCG AGC GTA GGC GTC GG 3′ and IS2-5′ CTC GTC CAG CGC CGC TTC GG 3′), and 0.5μl of Taq DNA Polymerase (500U) Invitrogen®, in a final volume of 50μl. The initial DNA denaturation occurred at 94°C for 2min, and after a total of 40 cycles of 94°C for 30s, 68°C for 2min, 71°C for 1min, and a final extension at 72°C for 10min1. The amplicons were revealed in 2% agarose gel, stained by ethidium bromide. The amplifications were developed blindly. Statistical analysis: the results of sensitivity and specificity, the positive and negative predictive values, and accuracy were compared with results of culture and phenotypic identification methods3,6. From the 87 smears with positive culture, 69 were from sputum, seven from bronchoalveolar lavage (BAL), six from tracheal aspiration, two from axillary abscesses, two from pleural liquids, and one from secretion (abscess); there was no sensitivity or specificity difference among these samples in the PCR amplification. The sensitivity and specificity of DNA amplification with IS6110 were 37.5 and 100%, respectively. The positive predictive value (PPV) was 100%, the negative predictive value (NPV) 80.8%, and accuracy was 82.8%. Among the 22 negative bacilloscopies with positive cultures of MTB, four were positive by PCR (18%). Among the 52 positive bacilloscopies, the sensitivity, specificity, PPV and NPV were 52.3%, 100%, 100% and 89.7%, respectively; accuracy was 90.7%. In Fig. 1, the electrophoresis of PCR IS6110 amplifications is demonstrated. The possibility of obtaining DNA from smears is a valuable alternative to solve the difficulties in transporting biological samples (long distances and biosecurity), and excludes the diagnosis of tuberculosis when the smear is positive due to the high specificity of IS6110, which is not possible when only stained microscopic preparations are used2.
Electrophoresis of IS6110 PCR amplifications. Line 1 – 100bp (bp=base pair(s) in DNA) DNA ladder; Line 2 – negative control; Line 3 – positive control – M. tuberculosis H37RV; Lines: 4, 6, 8, 9, and 10 – NTM samples; Lines: 5, 7, 11, and 13 – MTB samples; Lines: 14 and 15 – negative samples.
The high specificity found in this study is relevant, as it avoids the introduction of unnecessary treatments and conducts in the cases where bacilloscopy is positive and an NTM infection is in course. Therefore, this methodology could be applied in places where NTM species are often isolated, such as in Brazil, for instance, where during the period between 2000 and 2008, a total of NTM infection cases of 2139 were reported. Most of such cases were notified in the Southwestern region (1458), which includes the State of Minas Gerais, where this study was developed. The remaining cases out of this total were distributed among the Northern, Southern, Central-western, and Northeastern regions of the country11. The low sensitivity of in-house PCR has been associated with the small amount and heterogeneity of bacilli distribution in the samples; however, an increase in sensitivity could be observed when the bacilloscopies were positive, which is in accordance with prior reports in the literature11. Although in this study the IS6110 PCR was positive in only four samples among the negative bacilloscopies, this diagnosis was faster than the culture, which presented time for growth as a limiting factor2,7. The application of in-house IS6110 PCR in DNA extracted from smears coming from several samples (BAL, tracheal aspiration and secretions) is described herein for the first time, and because difference has not been observed in these results, this methodology has also applicative potential for TB diagnosis in other clinical specimens. The handling of clinical material during laboratory procedures, as well as its transportation, could generate higher risk for TB transmission. Hence, the possibility of using PCR in smears containing bacilli killed in the previous heat fixing by flame or heat plate decreases the risk related to such transmission, for the laboratory professionals who will conduct the DNA extraction. In order to handle the mycobacteria colonies, a more complex laboratory is necessary, requiring high maintenance investments as well as biosafety level III. Therefore, this methodology could be used in places without such a level of requirement, as is the case in several places in Brazil and all over the world; in addition, of course, to being an important tool for TB diagnosis or exclusion it when the smear is positive in cases of NTM infection6,12.
In conclusion, the IS6110 PCR in DNA extracted from a positive smear is a fast, simple, specific, and safe method.
Ethical disclosuresProtection of human and animal subjectsThe authors declare that no experiments were performed on humans or animals for this study.
Confidentiality of dataThe authors declare that no patient data appear in this article.
Right to privacy and informed consentThe authors declare that no patient data appear in this article.
Conflict of interestThe authors declare that they have no conflicts of interest.