A strain isolated from potato common scab superficial lesions in El Fuerte Valley in northern Sinaloa, Mexico, was identified by 16S rRNA and morphological methods. Moreover, the effects of the crude extract of strain V2 was evaluated on radish and potato. The isolate was similar to Streptomyces acidiscabies in its morphological properties; however, the 16S rRNA gene sequence of strain V2 was neither 100% identical to this species nor to the streptomycetes previously reported in Sinaloa, Mexico. Strain V2 did not amplify any specific PCR products for genes nec1 and tomA, which have been found and reported in S. acidiscabies. Strain V2 produced a PCR product for the txtAB operon, which is related to the production of thaxtomin. In vitro assays using crude thaxtomin extract and a spore suspension of the organism caused necrotic symptoms on radish and potato, which were highly virulent in potato. This study reports that Streptomyces sp. V2 has a toxigenic region (TR) that is associated with the thaxtomin gene cluster.
Se aisló una cepa de una lesión superficial de sarna común de la papa en un ejemplar procedente del Valle del Fuerte, en el norte de Sinaloa, México. La cepa fue identificada por secuenciación del gen 16S ARNr, y por sus características morfológicas. Los efectos del extracto crudo de dicha cepa, llamada V2, fue evaluado en papa y rábano. El aislado fue similar a Streptomyces acidiscabies en sus características morfológicas, pero la secuencia del gen 16S ARNr de la cepa V2 no fue 100% idéntica a la de dicha especie, ni tampoco a las de cepas identificadas dentro de este taxón previamente en Sinaloa, México. La cepa V2 no amplificó los productos específicos de PCR de los genes nec1 y tomA, los cuales sí se han reportado en S. acidiscabies. La cepa V2 amplificó el producto de PCR para del operón txtAB, relacionado con la producción de taxtomina. A través de ensayos in vitro usando un extracto crudo de taxtomina y una suspensión de esporas del organismo aislado se verificó la producción de síntomas necróticos en rábano y papa, con mayor virulencia en esta última especie. Este estudio indica que Streptomyces sp. V2 tiene una región toxigénica (TR) asociada con el cluster de genes de taxtomina.
The genus Streptomyces is best known mainly for its innate ability to produce biologically active secondary metabolites but also for the production of enzymes that degrade recalcitrant polymers such as cellulose, chitin, and lignin. Despite these biotechnologically important features, a small number of species are associated and/or related to economic losses in agriculture (e.g., as the causing agent of potato common scab [PCS]2,4,9,12,28,30–32,35) and to human disease (e.g., as the causing agent of actinomycetoma, a neglected chronic disease24). PCS is a disease that causes corky-like superficial necrotic lesions on potato that vary in color from brown to black and range in appearance from small raised tissue around the lenticel to large deep sunken pits up to 7mm in depth. The ability of PCS agents to cause damage in their hosts depends strongly on their ability to produce phytotoxins called thaxtomins3,15,23. Thaxtomins are cyclic peptides (2,5-diketopiperazines) resulting from the condensation of l-phenylalanine and L-4 nitrotryptophan. Up to 11 thaxtomins have been identified, with thaxtomin A being the most abundant of all3,15,23. The synthesis of thaxtomin A is associated with cell proliferation, expansion and necrosis when applied to immature periderm of potato tubers. Thaxtomin A is produced by Streptomycesacidiscabies, Streptomycesscabies (syn Streptomycesscabiei) and Streptomycesturgidiscabies, although other PCS-causing disease agents also synthesize minor quantities of this phytotoxin15. These pathogens possess a pathogenicity island (PAI) and the genes conferring pathogenicity are generally clustered. The PAI that appears to be responsible for the emergence of a new pathogenic species carries genes encoding four pathogenicity and virulence factors: (1) the biosynthetic pathway for thaxtomin A (ThxtA), (2) a functional tomatinase (TomA), (3) a small secreted protein (Nec1), and (4) a cytokinin biosynthetic pathway13. These four virulence factors also exist in S. scabiei and S. acidiscabies, but are separated in two remote chromosomal regions, designated as the toxicogenic region (TR) and the colonization region (CR)7,21,34. The genes associated with toxin production are clustered in the TR of the PAI7,21. S.acidiscabies is considered an emergent pathogen able to tolerate a lower pH and causing acid scab (AS), essentially the same as PCS except that it can occur in acidic pH soils where PCS is suppressed. Different reports of the AS causing agent show a worldwide distribution of the pathogen in northeastern US (first report), Canada, China, Germany, Japan, Korea, Norway and the UK4,9,12,19,32,35. Although the presence of S. acidiscabies is generally reported to occur in acidic soils (4.5–5.5)18, PCS caused by S. acidiscabies was recently reported in commercial potato fields at pH 7.0–7.2 in Sinaloa, Mexico26. The authors of the study also evaluated the pathogenicity of S. acidiscabies species in potato, radish, carrot and beet under greenhouse conditions at higher soil pH values (7.0–7.2). Other strategies such as species-specific methods have been used to detect pathogenic Streptomyces species from soil and potato tubers4. Phytotoxins, such as borrelidin and concanamycin, have also been reported to contribute to PCS, but none of these have been shown to be produced by the AS agent. Thus, cumulative evidence indicates that the pathogenic ability of S. acidiscabies to cause the disease relies solely on the thaxtomin A synthesis, and nec1 and tomA activities2,3,5,9,13,15,31. The objectives of this research were to identify a PCS streptomycete coded V2 and to evaluate the in vitro pathogenicity of its crude extract of thaxtomin compounds that has not been previously reported.
Materials and methodsStrain isolationStrain V2 was isolated from superficial PCS lesions collected in El Fuerte Valley, northern Sinaloa, Mexico (September 12th, 2009; Coordinates L 25.86895 and L −108.93087). The strain was recovered after scratching potato common scab lesions with a sterile cotton swab that was used to inoculate water agar plates supplemented with the antibiotics suggested by Williams and Davies33. Plates were incubated at 28°C for 15 days.
Organisms and culture conditionsOatmeal agar was prepared according to Shirling and Gottlieb's protocol and used as the routine media for strain V2 and S.acidiscabies ATCC 49003T27. The microorganisms were incubated at 28°C for 7 days; spore suspensions were maintained at 4°C and a biomass suspension in glycerol (20%, v/v) at −20°C prepared for long-term conservation as previously performed for other actinobacteria24.
Phenotypic characterizationPhenotypic characteristics of strain V2 and S. acidiscabies ATCC 49003T were evaluated by the methods described in the International Streptomyces Project (ISP) to observe colony morphology, melanoid and diffusible pigment production27.
16S rRNA gene amplificationAmplification of the 16S rRNA gene sequence was performed using universal primers 27f and 1525r as described previously with DNA extracted from strain V225. The PCR mixture was prepared in a total volume of 25μl: containing 0.5μl DNA, 2.5μl 10×-Buffer, 0.75μl 50mM MgCl2, 0.625μl 10mM dNTPs, 0.1μl Taq DNA polymerase (all Bioline, USA). PCR conditions were as follows: 10min at 95°C, 35 cycles with 1min at 95°C, 1min at 55°C, 1min at 72°C and one cycle of 10min at 72°C. An almost complete 16S rRNA gene sequence (≈1400pb, Macrogen, USA; PCR products of V2 were sequenced three times) was compared using the BLAST option in the NCBI database and a phylogenetic tree constructed using the Maximum Likelihood algorithm8, under the MEGA Software version 7.016, under the Kimura-2-parameter14. The stability of the resulting tree (constructed with the most related 16S rRNA gene sequences from the BLAST result) was assessed by performing a bootstrap analysis based on 1000 resamplings11.
Identification of genes associated with pathogenicityThe presence of predicted virulence-related genes was carried out by PCR using specific primers and PCR conditions for thaxtomin A and B (txtAB), tomatinase enzyme (tomA) and necrogenic protein (nec1) coding genes5,31. Primers and the expected size for the PCR product are indicated in Table 1.
Primers used for PCR detection of virulence-related genes
| Gene | Primer pair | Annealing T (°C) | Product size (pb) |
|---|---|---|---|
| nec1 | Nf: 5′-ATGAGCGCGAACGGAAGCCCCGGA-3′ | 60 | 700 |
| Nr: 5′-GCAGGTCGTCACGAAGGATCG-3′ | |||
| txtAB | TxtAB1: 5′-CCACCAGGACCTGCTCTTC-3′ | 48 | 385 |
| TxtAB2: 5′-TCGAGTGGACCTCACAGATG-3′ | |||
| tomA | Tom3: 5′-GAGGCGTTGGTGGAGTTCTA-3′ | 55 | 392 |
| Tom4: 5′TTGGGGTTGTACTCCTCGTC-3′ |
The method employed for this purpose was similar to that described by Leiner et al20. Briefly, 100ml of oatmeal broth (OMB) was inoculated with spore biomass of strain V2 and S. acidiscabies ATCC 49003T, as positive control. Strains were incubated at 28°C at 180rpm for 5 days in a rotatory shaker (New Brunswick, USA). Cultures were then centrifuged to remove both biomass and mycelium and the resulting filtrate was extracted twice with chloroform (1:1; Sigma–Aldrich, Mexico). After drying, the yellow residue was dissolved in methanol (Sigma–Aldrich, Mexico) and purified by passing the solution through a SUPELCO HPLC C18 column also in methanol. This crude extract was analyzed by reversed-phase thin-layer chromatography on MERCK HPTLC RP-18 (10×10cm) using acetone and water (3:2) as the solvent system. The same procedure was employed for S. acidiscabies ATCC 49003T and used for comparison purposes.
High-performance liquid chromatography (HPLC) was performed on an Agilent Technologies 1260 liquid chromatograph with a Zorbax SB C18 Agilent Technologies column (4μm, particle size, 150×4.6cm; Agilent Technologies Mexico S. R.L. de C.V.) using a water/acetonitrile 40:60 (v/v) mobile phase at a flow rate of 1ml/min. Ten microliters of crude extract were employed and the results monitored at 215 and 380nm.
In vitro pathogenicity assaysSlices of potato and radish were used to evaluate the phytotoxicity of crude extracts produced by strain V2. Separately, a spore suspension was also inoculated into potato slices to confirm the virulence of the isolate and evaluated after 72h. Streptomyces acidiscabies ATCC 49003T was used as a positive control as described previously and sterile distilled water as the negative control. Briefly, both potato tuber slices and radish seedlings were surface-disinfected with 1.0% NaOCl and 5×5cm slices then placed on moistened sterile filter paper in sterile Petri dishes after inoculation with either crude extract or spore suspensions (both 7μl) of each of the strains. Crude extract and spore suspensions were obtained by following the protocol proposed by Leiner et al20. All tuber slice experiments were repeated twice and incubated at 28°C in the dark.
ResultsPhenotypic characterizationStreptomyces sp. V2 produced a white spore mass color in ISP 1, 3, 4, 5, 6 and 7 media, however, no diffusible pigments were produced in ISP 3, 6 or 7 media. Morphological features of S. acidiscabies ATCC 49003T corresponded to those previously reported by Lambert and Loria18.
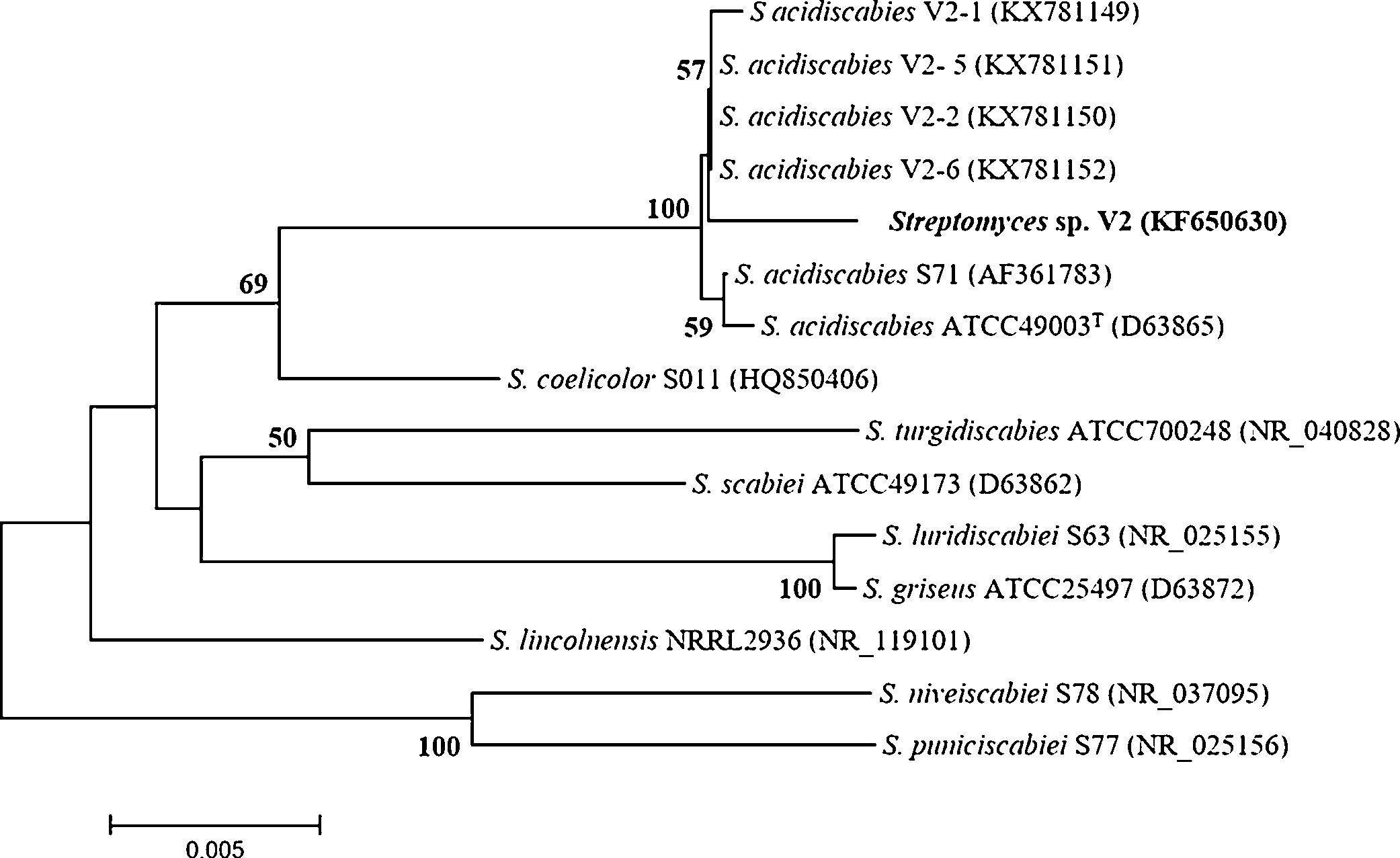
Phylogenetic identificationPutative assignment of strain V2 (Accession number KF650630) to the genus Streptomyces was straightforward after comparison against scabby streptomycete sequences from the GenBank database. The sequence was not 100% identical to those of the S. acidiscabies species (ATCC 49003T, V2-1, V2-2, V2-5 and V2-6) found in the database. The phylogenetic tree confirmed its assignment to the genus Streptomyces and revealed that it clustered with S. acidiscabies species (Fig. 1). The similarity values between strain V2 and S. acidiscabies ATCC 49003T were 99.4%, Streptomyces sp. V2 and V2-1, V2-2, V2-5 were 99.5% and Streptomyces sp. V2 and V2-6 were 99.6%, respectively.
Identification of genes associated with pathogenicityStrain V2 did not produce PCR products for two of the three pathogenic gene sets of primers tested as shown in Figure 2.. Strain V2 only yielded a PCR product for the txtAB set of primers whereas S. acidiscabies ATCC 49003T yielded the three expected PCR gene size products.
Extraction and chromatographic analysis of thaxtominStrain V2 produced the characteristic yellow compounds resembling thaxtomin when grown in OMB (oatmeal broth). Chromatographic analyses confirmed the production of thaxtomin compounds for strains V2 and S. acidiscabies ATCC 49003T. The TLC plate showed a total of five compounds (C1, C2, C3, C4 and C5) for S. acidiscabies ATCC 49003T and strain V2 (Fig. 3). The HPLC analysis identified four major peaks coming out of the extracts (Fig. 4). The most abundant compound produced by strain V2 was found at 1.8 (72%) and at 1.8min (52%) for S. acidiscabies ATCC 49003T (Table 2). The retention times of all peaks, areas and percentages are reported in Table 2.
Retention times, areas and percentages of thaxtomin compounds produced by the evaluated strains
| P | S. acidiscabies ATCC 49003T | StrainV2 | ||||
|---|---|---|---|---|---|---|
| RT | A | % | RT | A | % | |
| 1 | 1.240 | 5.81 | 38 | 1.227 | 3.40 | 18 |
| 2 | 1.693 | 0.84 | 5 | 1.733 | 1.03 | 5 |
| 3 | 1.887 | 7.89 | 52 | 1.873 | 13.60 | 72 |
| 4 | 2.053 | 0.76 | 5 | 2.053 | 1.02 | 5 |
Note: P, peak; RT, retention time; A, area in cm2;%, percentage.
The necrotic zone on slices of both potato and radish treated with crude extract and spore suspension of strain V2 were observed after 72h and similar to those produced by the positive control (S. acidiscabies ATCC 49003T). The damage on the vegetable tissue and the reported brown color from both essays is shown in Figure 5 (crude extract and spore suspension, respectively), thus confirming the ability of strain V2 to cause necrotic symptoms in in vitro pathogenic assays.
DiscussionStreptomycetes have always attracted attention due to their innate ability to produce secondary metabolites, most of them of biotechnological importance and extracellular enzymes, including cellulases, pectinases, among others6. However, there are few studies on pathogenic streptomycetes, and most of them focus on no more than 15 species causing either human or plant disease4,9,12,19,24,32,35. The symptoms of PCS in El Fuerte Valley, in the northern part of the State of Sinaloa (Mexico) differ from potato growing areas in the southern part of the state, where PCS is caused by S. scabiei and from reports from the USA26,32. It is important to firmly establish the identity of PCS-causing agents in order to appropriately design epidemiological and local studies that can lead to the successful prevention of PCS in particular regions in Mexico and elsewhere4,9,12,19,22,26,32,35. In this context, a species-specific method to detect pathogenic Streptomyces species from soil and potato tubers in Argentina has shown the presence of S. acidiscabies and S. turgidiscabies4. The combined PCR method showed notable advantages; however, it was also mentioned that further studies should isolate other pathogenic Streptomyces species4. The latter statement shows the importance of the isolation of pathogenic streptomycetes to carry out etiological, epidemiological and biogeographic studies to complement molecular methods10,29. It has been reported that the sequences of the 16S rRNA gene of S. acidiscabies ATCC 49003T and V2-1, S. acidiscabies V2-2, V2-5 and V2-6 were 100% identical26. However, the corresponding 16S rRNA gene sequence of V2 was not 100% identical to those of S. acidiscabies ATCC49003T, V2-1, V2-2, V2-5 and V2-6. The corresponding comparison of strain V2 against its closest phylogenetic relatives, that is, S. acidiscabies ATCC 49003T and V2-1, S. acidiscabies V2-2, V2-5 and V2-6, showed similarity percentages in the range of 99.4 to 99.6%. Although these percentages may suggest that strain V2 and the closest S. acidiscabies species are the same organism, other streptomycete species show higher similarity percentages and are distinct species, for instance, 99.7% between S. scabiei and Streptomyces europaeiscabiei. Therefore, there is a possibility that strain V2 is a distinct organism from S. acidiscabies ATCC49003T, though further taxonomic studies are certainly required. Indeed, genomic comparisons of strain V2 and S. acidiscabies genomes publicly available also suggest so1. Strain V2 seems to represent a novel species within the genus Streptomyces and is closely related to clade 5 within the family Streptomycetaceae17.
Strain V2 only amplified operon txtAB, whereas S. acidiscabies pathogenicity has been related to nec1, tomA and txtAB operons for which PCR primers have been designed and previously reported5,31. This was particularly interesting because it suggests that strain V2 may contain and/or express other genes that could also be related to PCS disease, opening a wide range of possibilities and lines of research to understand this phenomenon in the affected regions of Mexico. The necrotic symptoms observed in the in vitro tests with potato and radish were produced by the crude extract of thaxtomin, suggesting that necrosis is associated with the thaxtomin gene cluster of the toxicogenic region (TR)7,21,34. The spore suspension assay showed that strain V2 produced the necrotic symptoms of the disease and that V2 was 65% more virulent in potato compared to S. acidiscabies (25%). The thaxtomin extracts in both strains V2 and S. acidiscabies ATCC 49003T had similar retention times; however, they were present in higher amounts in the extract obtained from strain V2 when compared against S. acidiscabies ATCC 49003T.
The genetic analyses of strain V2 did not show the presence of other pathogenic markers expected and detected in S. acidiscabies species. This was an intriguing finding because both strains generated a similar pattern of crude-extract compounds by HPLC (Fig. 5).
Fyans and colleagues studied streptomycetes recovered from PCS in Newfoundland and reported some isolates related to S. europaescabiei; however, another group of isolates were not related to any plant-pathogenic Streptomyces12. Furthermore, these authors suggested that novel virulence factors may be contributing to the plant-pathogenic phenotype of these strains because of the unexpected variability of absence/presence of the already-known pathogenic markers in the evaluated strains. In our view, their independent but related work provides further evidence that there are indeed other putative novel species causing PCS not only in Canada but also – as we have proven in this study – in other regions of the world4,9,12,19,28,31,32,35. Comparative studies, both culture-dependent and culture-independent, among the most common PCS agents are certainly needed to tackle and extend our understanding of the disease worldwide4,9,12,19,22,26,29,32,35. Current studies are also on their way to properly describe strain V2 as another plant pathogenic streptomycete.
Conflict interestThe authors declare that they have no conflicts of interest.
AAV acknowledges a Ph.D. Scholarship 246635 from Consejo Nacional de Ciencia y Tecnología CONACYT, with international mobility at Universidad de Salamanca-USAL (CONACYT Becas Mixtas-2015) and Scholarship from Beca de Estímulo Institucional de Formación de Investigadores BEIFI-IPN (SIP-IPN 20130387-20160295). ETQ acknowledges IPN grants SIP 20160295, SIP20170432 and CONACYT C-291045.86/SIP-2016-RE/046. Authors are grateful to Prof. Rosemary Loria (Department of Plant Pathology, University of Florida, Gainesville USA) for providing S. acidiscabies.