This study was undertaken to investigate the resistance phenotypes to macrolide-lincosamide-streptogramin B (MLSB) antibiotics and their associated genotypes in isolates of Staphylococcus aureus. We analyzed one hundred, consecutive, non-duplicate isolates (methicillin-susceptible MSSA, n=53 and methicillin-resistant MRSA, n=47) obtained from various clinical samples between July 2012 to December 2013. The resistance profile to MLSB antibiotics was determined by phenotypic methods and the resistance genes were detected by PCR assays. All of the isolates were subjected to pulsed-field gel electrophoresis (SmaI-PFGE). The overall prevalence of resistance to MLSB antibiotics was 38% and the resistance phenotype distribution was as follows: cMLSB, 22%; iMLSB, 10%; MSB, 5% and L, 1%. We detected ermA, ermC, ermB and mrsA/B genes in these resistant isolates. The single ermA gene was commonly observed mainly in those with a cMLSB R phenotype, whereas the combination ermA and ermC was more commonly observed in isolates with inducible expression. The patterns of SmaI-PFGE suggest a great genetic diversity in both MRSA and MSSA resistant to MLSB antibiotics. The results demonstrate the local presence of S. aureus resistant to MLSB antibiotics and its most frequently described responsible genes. Some of these isolates, especially those with the iMLSB phenotype, may be associated with therapeutic failure. Therefore, efforts should be directed to the correct detection of all MLSB resistant isolates using appropriate laboratory tests. PFGE results reveal a wide spread of resistance genes rather than the circulation of S. aureus clones resistant to MLSB antibiotics.
Los objetivos de este estudio fueron investigar en Staphylococcus aureus la presencia de fenotipos resistentes a los antibióticos macrólidos, lincosamidas y estreptograminas tipoB (MLSB) y conocer sus genotipos responsables. Analizamos 100 aislamientos consecutivos, no duplicados (53 sensibles a meticilina [MSSA] y 47 resistentes a meticilina [MRSA]), obtenidos entre 2012 y 2013 a partir de diferentes muestras clínicas. El perfil de resistencia a los antibióticos MLSB fue determinado por métodos fenotípicos y los genes de resistencia se detectaron por PCR. Todos los aislamientos fueron comparados por SmaI-PFGE. La prevalencia global de resistencia a los antibióticos MLSB fue del 38% y la distribución de los fenotipos de resistencia fue la siguiente: cMLSB, 22%; iMLSB, 10%; MSB, 5%; L, 1%. Se detectaron los genes ermA, ermC y mrsA/B en los aislamientos resistentes. El gen ermA se observó, sobre todo, en aislamientos con fenotipo resistente constitutivoR (cMLSB), mientras que la combinación ermA y ermC se detectó principalmente en aislamientos con resistencia inducible (iMLSB). Los patrones de SmaI-PFGE sugieren una gran diversidad genética en los aislamientos resistentes a los antibióticos MLSB, tanto MRSA como MSSA. Los resultados demuestran la presencia local de S. aureus resistentes a los antibióticos MLSB y de sus genes responsables más frecuentemente descritos. Estos cultivos, especialmente aquellos con fenotipo resistente iMLSB, pueden asociarse con fallas terapéuticas. Por lo tanto, los esfuerzos deben dirigirse a la correcta detección de todos los cultivos resistentes a MLSB utilizando pruebas de laboratorio adecuadas. Los resultados de SmaI-PFGE sugieren una amplia diseminación de genes de resistencia, más que la circulación de clones resistentes a los antibióticos MLSB.
Staphylococcus aureus is a versatile microorganism that is found in the human microbiota and is also responsible for both hospital- or community-acquired illnesses. The diseases that it causes range from mild lesions that compromise skin and soft tissues to more severe conditions such as pneumonia, septic arthritis, endocarditis, osteomyelitis, and sepsis12,31,33. A biological characteristic of S. aureus is its ability to acquire exogenous genetic material that encodes several resistance mechanisms for different antibiotics. The mobile staphylococcal chromosomal cassette mec (SCCmec) that carries the mecA gene which encodes PBP2a, which is responsible for methicillin-resistance (MRSA), was likely acquired by horizontal transfer from a related species8. Three categories of MRSA are currently recognized, healthcare-associated (HA-MRSA), community-associated (CA-MRSA) and livestock-associated MRSA (LA-MRSA) according to epidemiological criteria, site of infection acquisition (community, hospital or animal contact), associated antibiotic susceptibility profile and genotype, including SCC element type11,34,35. S. aureus belonging to the three aforementioned categories have been implicated around the world in both sporadic cases and outbreaks of human staphylococcal diseases4,16,37. Macrolides, lincosamides, and type B streptogramin (MLSB) antibiotics can be alternatives for treatment of MRSA infections36. However, the increase in macrolide, lincosamide and type B streptogramin resistant isolates complicates the empirical antimicrobial treatment selection in S. aureus infections14. A fairly common resistance mechanism observed in S. aureus is due to the action of efflux pumps, encoded by the mrsA and mrsB genes responsible for pumping macrolide and streptogramin B antibiotics out of the bacteria (MSB resistance phenotype). On the other hand, the exclusive resistance to lincosamides (L resistance phenotype) mediated by lincomycin nucleotidyl-transferase enzymes and codified by genes lnuA-F, is occasionally observed. Isolates that carry lnu genes resist high levels of lincomycin but can be susceptible to clindamycin in the disk induction test18,23. In S. aureus and other gram-positive bacteria, resistance to MLSB antibiotics is mainly due to the 23S rRNA modification by adenine-N methyltransferase enzymes. This modification compromises macrolide, lincosamide and type B streptogramin activity. Genes encoding these methylases have been designated as erm genes and their expression can be constitutive (cMLSB resistance phenotype) or inducible (iMLSB resistance phenotype)13. Isolates with the iMLSB phenotype show in vitro resistance to erythromycin, but can falsely seem to be susceptible to clindamycin. These isolates have a high rate of spontaneous mutation to constitutive resistance phenotypes that can be selected by the usage of clindamycin, resulting in therapeutic failure. This fact highlights the importance of performing a correct laboratory identification using any of the recommended tests3,6. In our region, data about the prevalence of resistance to MLSB antibiotics in isolates of S. aureus recovered from adult individuals are scarce. This study aimed to investigate the presence of the different erythromycin and clindamycin resistance phenotypic profiles and also to determine the responsible genotypes in a set of S. aureus isolates recovered in a public, tertiary level hospital of Montevideo city, Uruguay.
Materials and methodsWe study 100 consecutive, non-duplicate S. aureus isolates obtained from patients admitted at the “Hospital Pasteur” (HP) from July 2012 to December2013. Setting The HP is a 250-bed tertiary level public and teaching hospital for all medical and surgical specialties. Patients receiving medical care at the HP are adults (>16 years of age) and often come from low-income households.
Bacterial isolates, source, identification and antimicrobial susceptibility testing.
S. aureus isolates were identified using the automated system VITEK 2 version 7.01 and classified as methicillin-resistant (MRSA) or methicillin-susceptible (MSSA) according to the obtained MIC value to oxacillin and the result of the cefoxitin test. The site of infection was taken from the laboratory records. Erythromycin (ERY) and clindamycin (CLI) phenotypes were assessed and interpreted using the D-zone test published by Steward et al.32 (D phenotype: D-shaped clear zone around the CLI disk near the ERY disk; D+ phenotype: like the previous one but with small colonies growing close to the CLI disk in an otherwise clear zone; HD phenotype: two zones of growth around the CLI disk: one zone is a light growth extending from the CLI disk to the second zone, where the growth is much thicker; R phenotype: growth to the edge of both CLI and ERY disks; MSB phenotype: a clear zone around the CLI disk; L phenotype: growth only up to the CLI disk; S phenotype: susceptible to both the CLI and ERY disks)3,2.
The IHSA 23 strain of S. aureus was used as a positive control for the D-test. Minimum inhibitory concentration (MIC) to erythromycin, clindamycin, vancomycin, teicoplanin and oxacillin was determined by the agar dilution method according to the Clinical and Laboratory Standards Institute recommendations6. The susceptibility patterns to ciprofloxacin, gentamicin, trimethoprim-sulfamethoxazole, tetracycline and rifampicin were taken from the VITEK report. Broth macrodilution assays (BMA) that included combinations of CLI plus ERY were conducted with minor modifications in accordance with the general procedure previously described by Steward et al.32. Briefly, the combinations were tested using CLI at final concentrations ranging from 0.5 to 8μg/ml plus ERY at final concentrations ranging from 0.03 to 0.12μg/ml. Reading was done after 18–24h of incubation at 35–37°C. This procedure was applied to those isolates that showed the iMLSB, cMLSB, MSB and S phenotypes by disk diffusion tests. S. aureus ATCC 29213 (CLI and ERY-susceptible) was included as control. The isolates were stored at −20°C in skimmed milk for further studies.
Molecular typing and antibiotic resistance gene detection. Bacterial DNA was obtained from isolated colonies using the Wizard genomic DNA preparation kit (Promega. Madison, Wis, USA) adding 20mg/ml lysostaphin (Sigma Chemical) in the cell lysis step. Detection of the mecA gene and SCCmec typing was performed in all MRSA isolates by a multiplex PCR assay previously described by Oliveira DC and H. de Lencastre20. MRSA isolates that carried the SCCmec type IV were arbitrarily taken as community-associated methicillin-resistant S. aureus (CA-MRSA) while those that carried the SCCmec type I, II, or III were grouped as health-care associated methicillin-resistant S. aureus (HA-MRSA). PCR detection of the ermA, ermB, ermC, msrA and msrB genes was performed on all isolates that showed the following resistance phenotypes: iMLSB (D and D+); cMLSB (HD and R); and MSB using standard protocols and previously described primers15. S. aureus MM122-35 was used as positive control for the ermA gene, S. aureus MM2 was used as a positive control for the ermB gene, and S. aureus MM523 for the ermC gene. S. aureus MM3627 was included as positive control for both msr A and B genes. All the strains were kindly provided by Genoveva Pensado (Bacteriology and Virology department). Ultrapure RNAse/DNAse free water was used as negative control in all PCR reactions.
Pulsed field gel electrophoresis (PFGE). DNA macro-restriction with the SmaI enzyme and further separation of fragments by pulsed field gel electrophoresis (PFGE) were carried out using a previously described protocol24. The obtained profiles were analyzed by the unweighted pair group method with arithmetic average (UPGMA) using BioNumerics v.7.1. The isolates that showed ≥80% similarity were included into the same pulsogroup (PG), while those that showed ≥95% similarity were considered to be of identical pulsotype (PT)9. S. aureus subsp. aureus (strain NCTC 8325) was included as reference size marker.
ResultsSource of analyzed S. aureus isolates and resistance phenotypesWe studied 100 isolates of S. aureus. Twenty-three of them were recovered from invasive diseases including bacteremia (n=10; 43.4%), ventilator-associated pneumonia (n=8; 35%), deep abscesses (n=4; 17.3%) and acute meningoencephalitis (n=1; 4.3%). On the other hand, 77 isolates were recovered from skin and soft tissue infections (SSTIs) including superficial abscesses (n=39; 50%), wounds (n=22; 28.5%), cellulitis (n=11; 14.2%) and whitlows (n=5; 6.4%).
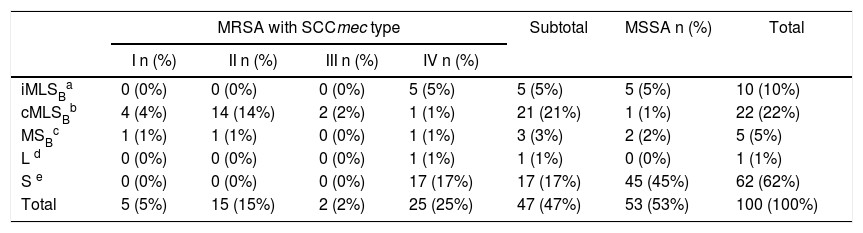
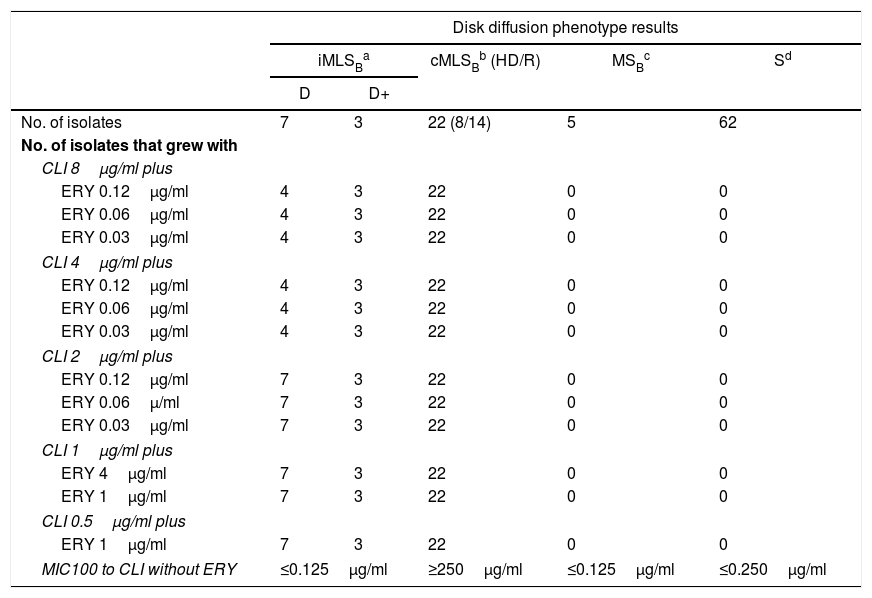
Fifty-three isolates were classified as MSSA and the remaining 47 as MRSA (all of them carried the mecA gene); 25 corresponded to CA-MRSA and 22 to HA-MRSA, according to the above described abbreviated criterion. The distribution of the SCCmec types in HA-MRSA was as follows: SCCmec type I, 5 isolates; SCCmec type II, 15 isolates and SCCmec type III, 2 isolates. Fifteen percent of the MSSA isolates were resistant to MLSB antibiotics, whereas this figure increased to 63.8% in the MRSA (CA-MRSA and HA-MRSA) isolates. All S. aureus isolates included in this study were susceptible to vancomycin and teicoplanin. Four isolates (4%) were resistant to rifampicin, all of them carrying the SCCmec type II. Twenty two out of 47 (46.8%) MRSA showed resistance to ciprofloxacin (HA-MRSA, n=21; CA-MRSA, n=1). However, only 4 out of 53 (7.5%) MSSA were resistant to this antibiotic. Eight S. aureus isolates (8%) showed resistance to gentamicin, 4 (7.5%) corresponded to MSSA isolates and 4 (8.5%) to MRSA (HA-MRSA, n=3; CA-MRSA, n=1). One methicillin-susceptible isolate was resistant to tetracycline. The only S. aureus isolate that showed resistance to trimethoprim-sulfamethoxazole belonged to the HA-MRSA group. Table 1 shows the resistant phenotypes to MLSB antibiotics found in MRSA and MSSA isolates. Table 2 shows the percentage of resistance to macrolides and lincosamides found in MRSA (HA-MRSA, CA-MRSA) and MSSA isolates. Three out of the 10 isolates with iMLSB resistance showed the D+ phenotype in the disk-diffusion test. The isolates with the iMLSB resistance phenotype (n=10; 5 CA-MRSA and 5 MSSA) showed MICs for CLI ≤0.125μg/ml and those with phenotype S (n=62; MSSA 45 and CA-MRSA 17) had MICs for CLI ≤0.25μg/ml. In those isolates that exhibited the cMLSB resistance phenotype (n=22; HA-MRSA, 21 and MSSA, 1 isolate) MICs for CLI were ≥256μg/ml. As shown in Table 3, the combination of 2μg/ml of CLI plus 0.12μg/ml of ERY in the BM assay allowed us to differentiate between isolates displaying iMLSB or cMLSB resistance phenotypes and those with the MSB or S phenotype. Seven out of 10 isolates that showed the iMLSB resistant phenotype (D, 4 isolates and D+, 3 isolates) grew in the presence of CLY at 8μg/ml plus ERY 0.12μg/ml and all the isolates (n=22) that showed the cMLSB resistance phenotype (HD and R) grew in the presence of the combination 8μg/ml of CLI plus ERY 0.12μg/ml.
Distribution of phenotypes to MLSB antibiotics in S. aureus isolates using the D-zone test.
| MRSA with SCCmec type | Subtotal | MSSA n (%) | Total | ||||
|---|---|---|---|---|---|---|---|
| I n (%) | II n (%) | III n (%) | IV n (%) | ||||
| iMLSBa | 0 (0%) | 0 (0%) | 0 (0%) | 5 (5%) | 5 (5%) | 5 (5%) | 10 (10%) |
| cMLSBb | 4 (4%) | 14 (14%) | 2 (2%) | 1 (1%) | 21 (21%) | 1 (1%) | 22 (22%) |
| MSBc | 1 (1%) | 1 (1%) | 0 (0%) | 1 (1%) | 3 (3%) | 2 (2%) | 5 (5%) |
| L d | 0 (0%) | 0 (0%) | 0 (0%) | 1 (1%) | 1 (1%) | 0 (0%) | 1 (1%) |
| S e | 0 (0%) | 0 (0%) | 0 (0%) | 17 (17%) | 17 (17%) | 45 (45%) | 62 (62%) |
| Total | 5 (5%) | 15 (15%) | 2 (2%) | 25 (25%) | 47 (47%) | 53 (53%) | 100 (100%) |
Results of Broth Macrodilution Assays (BMA).
| Disk diffusion phenotype results | |||||
|---|---|---|---|---|---|
| iMLSBa | cMLSBb (HD/R) | MSBc | Sd | ||
| D | D+ | ||||
| No. of isolates | 7 | 3 | 22 (8/14) | 5 | 62 |
| No. of isolates that grew with | |||||
| CLI 8μg/ml plus | |||||
| ERY 0.12μg/ml | 4 | 3 | 22 | 0 | 0 |
| ERY 0.06μg/ml | 4 | 3 | 22 | 0 | 0 |
| ERY 0.03μg/ml | 4 | 3 | 22 | 0 | 0 |
| CLI 4μg/ml plus | |||||
| ERY 0.12μg/ml | 4 | 3 | 22 | 0 | 0 |
| ERY 0.06μg/ml | 4 | 3 | 22 | 0 | 0 |
| ERY 0.03μg/ml | 4 | 3 | 22 | 0 | 0 |
| CLI 2μg/ml plus | |||||
| ERY 0.12μg/ml | 7 | 3 | 22 | 0 | 0 |
| ERY 0.06μ/ml | 7 | 3 | 22 | 0 | 0 |
| ERY 0.03μg/ml | 7 | 3 | 22 | 0 | 0 |
| CLI 1μg/ml plus | |||||
| ERY 4μg/ml | 7 | 3 | 22 | 0 | 0 |
| ERY 1μg/ml | 7 | 3 | 22 | 0 | 0 |
| CLI 0.5μg/ml plus | |||||
| ERY 1μg/ml | 7 | 3 | 22 | 0 | 0 |
| MIC100 to CLI without ERY | ≤0.125μg/ml | ≥250μg/ml | ≤0.125μg/ml | ≤0.250μg/ml | |
Inducible resistance to MLSB antibiotics, D: clear zone around the CLI disk near to the ERY disk; D+: like the previous one but with small colonies growing up to the CLI disk in an otherwise clear zone.
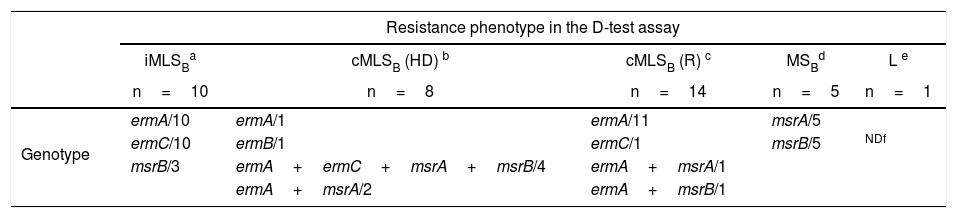
As shown in Table 4, all the isolates that showed the iMLSB resistance phenotype in the D-test carried the combination of the ermA and ermC genes. Instead, in those isolates that displayed the cMLSB resistance phenotype (HD or R) the predominant genotype was ermA (alone or in combination with other resistance genes). In those isolates that showed the cMLSB (HD) phenotype, the combination of the ermA and mrs genes was seen more frequently (6 out of 8) than in those that displayed the cMLSB (R) phenotype (2 in 14) (Table 4). The ermB gene was found in just one isolate with the cMLSB resistance phenotype. The responsible genotype was not studied in the CA-MRSA isolate that showed the L resistance phenotype.
Genotypes found in isolates of S. aureus resistant to MLSB antibiotics.
| Resistance phenotype in the D-test assay | |||||
|---|---|---|---|---|---|
| iMLSBa | cMLSB (HD) b | cMLSB (R) c | MSBd | L e | |
| n=10 | n=8 | n=14 | n=5 | n=1 | |
| Genotype | ermA/10 | ermA/1 | ermA/11 | msrA/5 | |
| ermC/10 | ermB/1 | ermC/1 | msrB/5 | NDf | |
| msrB/3 | ermA+ermC+msrA+msrB/4 | ermA+msrA/1 | |||
| ermA+msrA/2 | ermA+msrB/1 | ||||
cMLSB (HD): constitutive resistance to MLSB antibiotics (two zones of growth appear around the CLI disk, one zone is a light growth extending from the CLI disk to the second zone where the growth is much thicker).
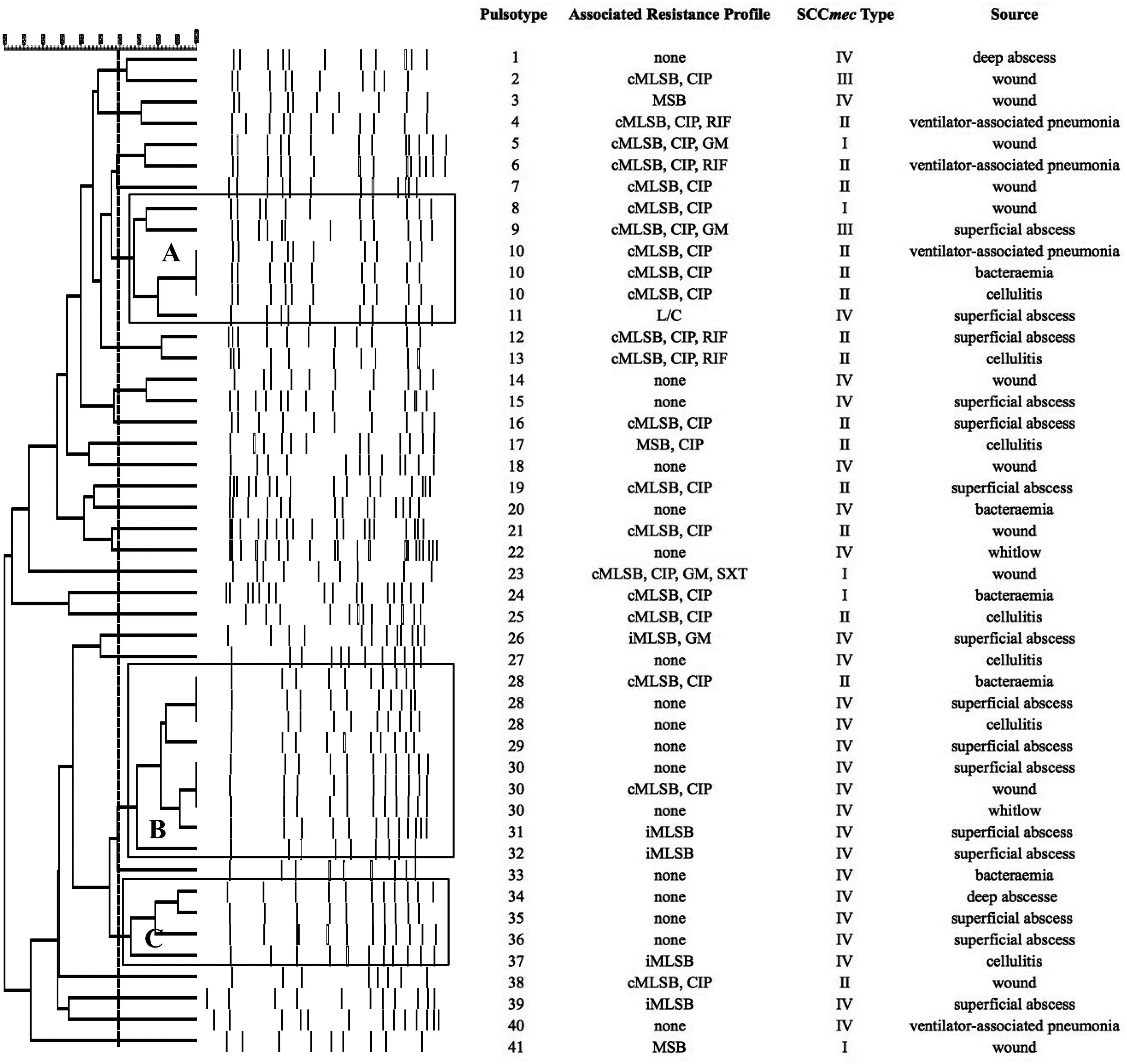
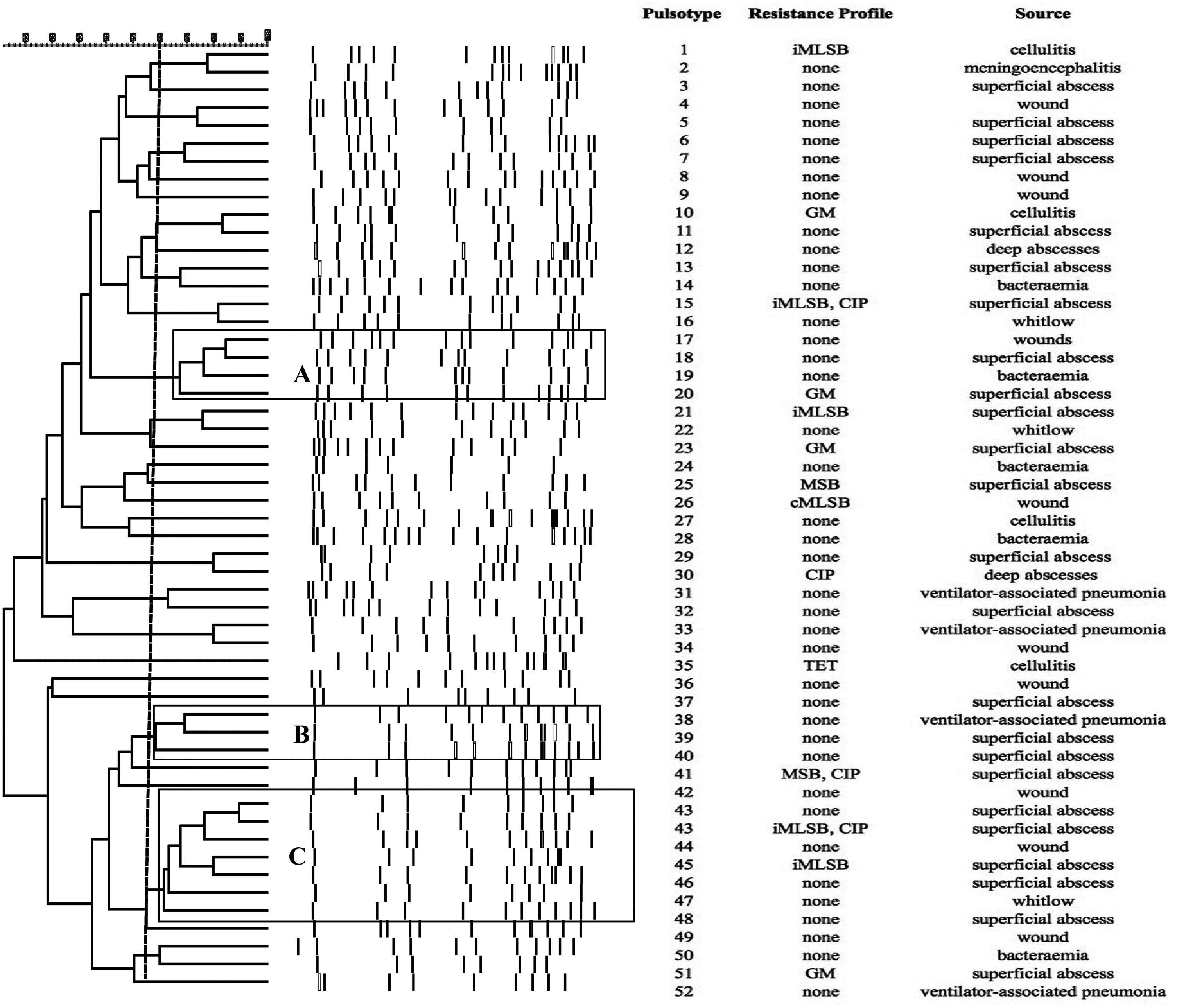
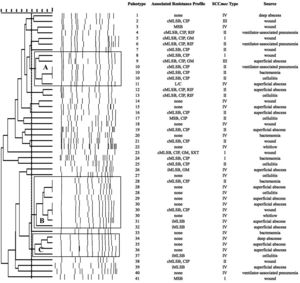
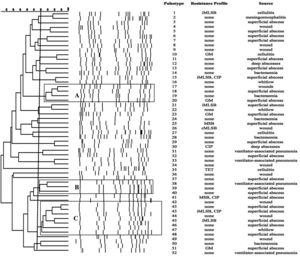
SmaI-PFGE profiles in MRSA and MSSA isolates. As shown in Figure 1, the SmaI-PFGE profile analysis of MRSA isolates revealed a wide variety of PTs (n=41) and 3 clusters (pulsogroups, PG: A, B and C, with ≥80% similarity and more than 2 isolates in each). Cluster A included 6 isolates, one with SCCmec IV, one with SCCmec III, one with SCCmec I and finally 3 isolates that carried the SCCmec II element with the cMLSB resistance phenotype that showed 100% identity and were recovered from 3 inpatients located in the same surgery ward. The only isolate with the L resistance phenotype recovered in this study was located in this PG. Cluster B (9 isolates) included 8 CA-MRSA isolates. Three of them (including an isolate carrying the SCCmec type II) showed 100% similarity and were visually identical to the main pulsotype found in local CA-MRSA isolates recovered in Uruguay between 2001 and 2002 (PFGE profile A1, type USA1100). Other 3 isolates in this PG showed 100% similarity among them (including the single CA-MRSA isolate with the cMLSB resistance phenotype recovered in this study) and were also visually similar to the CA-MRSA PFGE profile A1. Cluster C also included CA-MRSA isolates. The 5 CA-MRSA isolates that displayed the iMLSB resistance phenotype belonged to 5 different PTs (see Fig. 1). In the MSSA isolates, 52 PTs and 3 PGs (A, B and C, ≥80% similarity, more than 2 isolates in each) (see Fig. 2) were recognized. Only 2 isolates showed 95% similarity and were located in the PG C. This PG included 7 isolates, 2 of them with the iMLSB phenotype (see Fig. 2).
Pulsed-field gel electrophoresis (PFGE) dendrogram of methicillin-resistant S. aureus isolates showing PTs, PGs, associated profile of resistance, SCCmec type and sample source. Dashed line indicates PGs (named as A, B and C, with ≥80% similarity and more than 2 isolates in each). cMLSB, constitutive resistance to macrolide, lincosamide and type B streptogramin; iMLSB, inducible resistance to macrolide, lincosamide and type B streptogramin; MSB, resistance to macrolide and streptogramin B; L, resistance to lincosamides; CIP, resistance to ciprofloxacin; GM, resistance to gentamicin; RIF, resistance to rifampicin; SXT, resistance to trimethoprim-sulfamethoxazole; None, susceptible to all tested antibiotics.
Pulsed-field gel electrophoresis (PFGE) dendrogram of methicillin-susceptible S. aureus isolates showing PTs, PGs, associated profile of resistance and sample source. Dashed line indicates PGs (named as A, B and C, with ≥80% similarity and more than 2 isolates in each). cMLSB, constitutive resistance to macrolide, lincosamide and type B streptogramin; iMLSB, inducible resistance to macrolide, lincosamide and type B streptogramin; MSB, resistance to macrolide and streptogramin B; L, resistance to lincosamides; CIP, resistance to ciprofloxacin; GM, resistance to gentamicin; RIF, resistance to rifampicin; TET, resistance to tetracycline; None, susceptible to all tested antibiotics.
Clindamycin has been used for the treatment of severe staphylococcal diseases, taking into account some of its pharmacological properties. However, resistance can develop in isolates with the iMLSB phenotype, and spontaneous constitutively resistant mutants have arisen from such isolates during clindamycin therapy, determining therapeutic failures in some patients10. This is the first local report about the distribution of resistance phenotypes to MLSB antibiotics and their responsible genes in a set of S. aureus isolates recovered from adult inpatients in a public hospital. The overall prevalence of resistance to MLSB antibiotics was 38%. This figure is similar to that reported by Abbas et al.1 (40%) in a study conducted in India that included 500 isolates of S. aureus recovered in a tertiary hospital and otherwise higher than that also found in India by Prabhu et al. (28.42%)25. Similarly to what happened in Serbia and India, in the current study most S. aureus isolates (62%) showed susceptibility to both macrolide and lincosamide antibiotics. However, the distribution of the resistance phenotypes was different from that reported by those authors. In Serbia, iMLSB was the most frequent resistance phenotype detected (31.5–33.7%); instead in India it was MSB (17%) followed by both cMLSB and iMLSB resistance phenotypes (11.6% and 10.8%, respectively)21,25. In this study the overall data show that the predominant resistance phenotype was cMLSB (22%) followed by iMLSB (10%), MSB (5%) and L (1%). Similar figures were reported by Sedaghat in the Iranian teaching hospital and also by Paz Pereira et al. in neighboring Brazil3,28. The cMLSB resistance phenotype prevailed among HA-MRSA isolates (SCCmec I, II and III) (20/22; 91%), which is in concordance with similar studies carried out in other countries2,5,29,39. In this work the prevalence of the iMLSB resistance phenotype was similar among MRSA (CA-MRSA and HA-MRSA) (5/47; 10.6%) and MSSA (5/53; 9%) isolates. This result is not in concordance with the findings reported by Abbas et al.1 or Rahbar et al.26, which showed a higher prevalence of the iMLSB resistance phenotype in MSSA isolates. Furthermore, in contrast to the findings of Mišic et al.21 in Serbia, none of the HA-MRSA (SCCmec I, II and III) isolates studied here showed the iMLSB resistance phenotype (Table 1). The prevalence of MSSA isolates with the iMLSB resistance phenotype found in our study (9%) was lower than that reported by Shoji et al. in Japan (25.4%)30. As we will comment below, the results obtained by SmaI-PFGE showed great genetic diversity in both MSSA and MRSA isolates with iMLSB and cMLSB resistance phenotypes (Figs. 1 and 2). This suggests a broad diffusion of the responsible resistance genes rather than the circulation of a predominant PT. The overall prevalence of the MSB resistance phenotype was 5%; this figure being lower than that reported by Mišic et al.21 in Serbia (17.6%) but similar to the results (6%) obtained by Sedaghat et al.28 in the Iranian teaching hospitals21,28. The combination of msrA and msrB genes was detected in all (5) isolates that showed this resistance phenotype. Only one isolate showed the L resistance phenotype; as formerly seen in Argentina, which corresponded to a CA-MRSA isolate carrying SCCmec type IV18. The occurrence of this resistance type remains low at the HP hospital (1%) as was described previously17,21,23. The prevalence of resistance phenotypes to MLSB antibiotics vary depending on the analyzed country, geographic area and even the type of patient (inpatients vs. outpatients; hospital vs. community origin; children vs. adults; public vs. private institutions; patients or healthcare workers included, among others). This global trend highlights the need for each country, region or hospital to use its own bacterial resistance surveillance program2,5,26,29. The obtained results in the BM assay combining 2μg/ml of CLI plus 0.12μg/ml of ERY allowed us to differentiate between isolates displaying iMLSB or cMLSB phenotypes and those with the MSB or the S phenotype. However, in contrast with Steward et al.’s findings32, in this study some S. aureus isolates with an iMLSB resistance phenotype and positive D test were able to grow at higher concentrations of clindamycin. This could be partly explained by the fact that these isolates with the D or D+ phenotype carry the combination of erm and msr genes, as we discussed further32. All analyzed S. aureus isolates (n=100) were susceptible to vancomycin and teicoplanin. However, it is necessary to conduct an active surveillance to early detect the presence of glycopeptide-resistant strains. As expected, most of the S. aureus isolates grouped in this study as HA-MRSA (21 of 22) showed ciprofloxacin resistance. Only 4 of 100 isolates were resistant to rifampicin and all of them corresponded to HA-MRSA.
Resistance genes to MLSB antibiotics and genetic comparison by SmaI-PFGEThe PCR analysis identified the ermA gene as the most frequent MLSB resistance gene followed by ermC. The ermA gene was detected individually or in combination with other genes such as ermC, msrA and msrB in 30 (79%) out of 38 S. aureus resistant to MLSB antibiotics. In 12 of them, especially in HA-MRSA (11/14), it was individually detected and all these isolates showed a cMLSB resistance phenotype (R) (see Table 4). This result is in concordance with previous findings obtained in European hospitals27. In general, the ermA genes are mainly detected in MRSA isolates and are located in transposons related to Tn554, while the ermC genes are frequently responsible for resistance in MSSA and are located in plasmids. Based on the SmaI PFGE results, we can rule out the hypothesis of a broad hospital spread of a single pulsotype with a cMLSB resistance phenotype carrying the ermA gene. Only three isolates belonging to cluster A (Fig. 1) showed 100% similarity; they carried the SCCmec type II and were recovered from three patients of the same ward. This probably corresponded to a case of limited intrahospital cross contamination. In the two above cited studies, there are no data about the genetic comparison of the included strains; therefore, it cannot be ruled out that they pertained to prevalent clones. In similar studies conducted in Brazil and European countries the ermC gene predominated over the ermA gene19,22,37. As it occurred in Brazil, we merely detected one resistant isolate that carried the ermB gene and showed the cMLSB resistance phenotype7. This low prevalence (2.6%) was expected, taking into account that we only included isolates collected from adult patients. This gene is frequently found in S. aureus isolates from animal origin and also in pediatric CA-MRSA isolates7,38. The PCR analysis identified the msrA and msrB genes, alone or combined, in 16 isolates belonging to the MSSA, CA-MRSA or HA-MRSA categories with iMLSB, cMLSB and MSB resistance phenotypes. An interesting result of this study was the high rate of combination of MLSB resistance genes. The most commonly detected profile was ermA plus ermC (n=14), followed by the combination of ermA plus msrA (n=7), many of which also bore the msrB gene. All isolates with the iMLSB resistance phenotype (n=10) carried both ermA and ermC genes, and three of them also harbored the msrB gene (Table 4). The SmaI-PFGE results showed great genetic diversity in both MSSA and MRSA isolates with iMLSB and cMLSB resistance phenotypes. They support the wide spread of the resistance genes to MLSB antibiotics in these studied clinical S. aureus isolates rather than the circulation of a predominant PT. This study has two limitations: MRSA isolates were considered to be CA-MRSA or HA-MRSA only according to the SCCmec type that they carried and the use of PCR for erm genes does not allow to differentiate between isolates with iMLSB or cMLSB resistance phenotypes.
ConclusionsThe overall prevalence of resistance to MLSB antibiotics in this group of clinical S. aureus isolates was 38% and the resistance phenotype distribution was as follows: cMLSB, 22%; iMLSB, 10%; MSB, 5% and L, 1%. In these resistant isolates we detected, ermA, ermC, ermB and mrsA/B genes. The single ermA gene was commonly found in HA-MRSA (SCCmec I, II and III) isolates, mainly in those with a cMLSB R resistance phenotype; whereas the combination ermA and ermC was more commonly observed in MSSA or CA-MRSA (SCCmec IV) isolates with inducible resistance expression (iMLSB phenotype). There was a high rate of combination of MLSB resistance genes and the obtained patterns by SmaI-PFGE suggest a great genetic diversity in this group of MRSA and MSSA MLSB resistant isolates. The obtained results demonstrate the local presence of S. aureus resistant to MLSB antibiotics and its most frequently described responsible genes. These isolates (especially those with the inducible resistance phenotype, iMLSB) may be associated with therapeutic failures; therefore, efforts should be directed toward their correct detection using appropriate laboratory tests, particularly among those recovered from hospitalized individuals suffering from comorbidities or receiving immunosuppressive treatment.
FundingThis work was supported by a grant of the Comisión Sectorial de Investigación Científica (CSIC), Universidad de la República [program I+D group 290].
Conflict of interestThe authors declare that they have no conflict of interest.
We thank Dr. Felipe Schelotto for the critical review of the manuscript.