The aim of this study was to characterize phenotypically and genotypically 27 mecA positive Staphylococcus aureus strains with oxacillin MICs of ≤2μg/ml by Vitek 2, isolated in different regions of Uruguay. Susceptibility to oxacillin and cefoxitin was studied by gradient diffusion, disk diffusion to cefoxitin, and Phoenix and MicroScan systems. PBP2a was determined. SCCmec typing was performed and the isolates were compared by PFGE. Twenty-six isolates were susceptible to oxacillin; one strain was susceptible to cefoxitin by disk diffusion and 3 strains by gradient diffusion. Phoenix and MicroScan panels detected methicillin resistance in 25 and 27 strains, respectively. Twenty-six strains tested positive for PBP2a. Twenty-six strains carried SCCmec V and 24 belonged to pulsotype A. One strain carried SCCmec IV and did not belong to pulsotype A. Cefoxitin disk diffusion test and PBP2a detection correctly identified 26 of these 27 strains as MRSA. PFGE results suggest the dissemination of a cluster of MRSA carrying SCCmec V.
El objetivo de este estudio fue caracterizar fenotípicamente y genotípicamente 27 cepas de Staphylococcus aureus positivas para mecA y con CIM de oxacilina ≤2μg/ml según Vitek 2, obtenidas en diferentes regiones del país. La sensibilidad frente a la oxacilina y la cefoxitina se estudió por difusión en gradiente, por disco-difusión (cefoxitina) y por los sistemas Phoenix y MicroScan. Se analizó la portación de PBP2a, se realizó la tipificación de SCCmec y las cepas se compararon mediante PFGE. Resultaron sensibles a oxacilina por difusión en gradiente 26 cepas; una fue sensible a cefoxitina por disco-difusión y 3 lo fueron por difusión en gradiente. Los sistemas Phoenix y MicroScan detectaron resistencia a meticilina en 25 y 27 cepas, respectivamente. Asimismo, 26 cepas portaban PBP2a y 26 cepas mostraron presencia de SCCmecV, 24 correspondieron al pulsotipo A. Una portaba SCCmecIV y no perteneció al pulsotipo A. La prueba de disco-difusión con cefoxitina y la detección de PBP2a identificaron 26 de 27 cepas como MRSA. La PFGE sugiere la diseminación de un grupo MRSA con SCCmecV.
Staphylococcus aureus is responsible for a wide range of both community- and healthcare-acquired human diseases. It is a main cause of skin and soft tissue infections but can also produce life-threatening illnesses. β-lactam antibiotics are widely used in medicine being generally effective for the treatment of illnesses caused by this bacterium. However, there are methicillin-resistant S. aureus (MRSA) strains circulating around the world, questioning the empirical use of these antibiotics.15
Methicillin resistance in S. aureus is mostly mediated by the expression of an additional penicillin-binding-protein 2a (PBP2a) with low affinity for β-lactam antibiotics (except ceftobiprole and ceftaroline), encoded by the mecA gene or less frequently by the mecC gene. This fact determines oxacillin minimum inhibitory concentrations (MICs) of ≥4μg/ml. However, S. aureus carrying the mecA gene but oxacillin-susceptible (OS-MRSA) (MICs ≤2μg/ml) has been described worldwide, challenging its detection and treatment.1,3,11,13 The difference between the phenotypic and genotypic results could be explained by two previously known phenomena: the existence of strains with inducible resistance and strains with high heteroresistance to oxacillin. In the former case, S. aureus carrying the mecA gene becomes phenotypically oxacillin-resistant after exposure to oxacillin or cefoxitin; in the second case, gene expression and, therefore, oxacillin resistance, occur in a few cells in the original bacterial population that are selected when the entire population is exposed to the antibiotic.14 Furthermore, nucleotide changes within the mecA gene, determining the emergence of a premature stop codons that reverted by point mutations after exposure to cefoxitin, restoring the gene expression and therefore the resistant phenotype,7 were described in OS-MRSA mecA positive and PBP2a negative strains (known as “stealth methicillin-resistant S. aureus”).
In Uruguay, an OS-MRSA strain was previously reported in a case of infantile septic arthritis.6
Therefore, we set out to investigate whether other OS-MRSA strains were circulating in our country and to know their phenotypic and genotypic characteristics.
Between January and June 2018, we analyzed a set of 27, consecutive, non-duplicate S. aureus strains with oxacillin MIC values between 1 and 2μg/ml and negative cefoxitin screening test according to the Vitek 2 compact system (bioMérieux, AST-P577 card), software version 6.2.
The strains were isolated in different clinical microbiology laboratories at public and private health centers in Montevideo (n=19) and 3 other regions: Treinta y Tres (n=2), Durazno (n=2) and Canelones (n=4) and were recovered from skin and soft tissue infections (n=21), blood (n=4), joint fluid (n=1) and catheter tip (n=1). The strains were submitted to the Bacteriology and Virology department, Medicine School, Montevideo, Uruguay.
This work was approved by the Ethics Committee of the Centro Hospitalario “Pereira Rossell”.
We performed the cefoxitin disk diffusion test. Testing for oxacillin and cefoxitin was also done by gradient diffusion tests. Oxacillin MIC values of ≤2μg/ml and cefoxitin values of ≤4μg/ml were defined as susceptible. S. aureus ATCC 25923 and 29213 strains were used as quality controls 2.
The 27 strains were again analyzed by the Vitek 2 compact system (bioMérieux, AST-P577 card), software version 6.2, and also by Phoenix (Becton Dickinson) and MicroScan (Siemens Healthcare Diagnostics) panels.
PBP2a production was determined by the PBP2a SA culture colony test (Alere Inc., Scarborough, ME).
Detection of the mecA gene was carried out by PCR using the primers MECA P4 and MECA P7 according to Milheiriço et al.10 SCCmec typing was done by a multiplex PCR assay.9
The obtained profiles by pulsed field gel electrophoresis (PFGE) with SmaI enzyme were analyzed by the unweighted pair group method with arithmetic average (UPGMA) using BioNumerics v.7.1. The isolates with ≥80% similarity were included in the same pulsotype (PT), while those that showed ≥95% similarity were considered to have identical pulsotype.8S. aureus subsp. aureus (strain NCTC 8325) was included as reference.
In the disk diffusion test, 26 strains showed a cefoxitin inhibition zone of ≤21mm (resistant phenotype) whereas only one displayed a diameter of 22mm (susceptible phenotype) (Table 1). The mean value of the cefoxitin inhibition zone was 19mm (range 12–22).
Obtained results by different laboratory procedures in 27 OS-MRSA strain isolates in four Uruguayan regions. January to June 2018.
| Strain number | Laboratory procedure used | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Vitek* | BD Phoenix mg/l | MicroScan | Gradient diffusion mg/l** | Disk diffusion to cefoxitin*** | PBP2a testa | SCCmec typeb | |||||
| Oxacillin MIC | Cefoxitin Screening | Oxacillin MIC | Cefoxitin MIC | Oxacillin MIC | Cefoxitin Screening | Oxacillin MIC | Cefoxitin MIC | ||||
| 1 | 2 | − | 2 | >4 | 2 | + | 2 | 8 | 19 | + | V |
| 2 | 2 | − | 1 | 4 | 2 | + | 2 | 6 | 18 | + | V |
| 3 | 1 | − | ≤0.25 | 2 | 2 | + | 1 | 6 | 19 | + | V |
| 4 | 2 | − | 2 | >4 | 2 | + | 2 | 6 | 18 | + | V |
| 5 | 1 | − | 2 | >4 | 2 | + | 2 | 6 | 18 | + | V |
| 6 | 1 | − | 2 | >4 | 2 | + | 1.5 | 6 | 19 | + | V |
| 7 | 1 | − | 2 | >4 | 1 | + | 1.5 | 4 | 22 | + | V |
| 8 | 1 | − | 2 | >4 | 2 | + | 1 | 6 | 19 | + | V |
| 9 | 1 | − | 2 | >4 | 2 | + | 2 | 6 | 18 | + | V |
| 10 | 1 | − | 2 | >4 | 2 | + | 1.5 | 8 | 19 | + | V |
| 11 | 1 | − | 2 | >4 | 2 | + | 1.5 | 4 | 21 | + | V |
| 12 | 2 | − | 2 | >4 | 2 | + | 2 | 6 | 18 | + | V |
| 13 | 2 | − | 2 | >4 | 2 | + | 2 | 6 | 18 | + | V |
| 14 | 1 | − | 2 | >4 | 2 | + | 1.5 | 6 | 20 | + | V |
| 15 | 1 | − | 1 | >4 | 1 | + | 1.5 | 4 | 19 | + | V |
| 16 | 2 | − | 2 | >4 | 2 | + | 1 | 6 | 16 | + | V |
| 17 | 2 | − | 2 | >4 | 1 | + | 3 | 6 | 17 | + | V |
| 18 | 2 | − | 2 | >4 | 2 | + | 4 | 6 | 21 | + | V |
| 19 | 2 | − | 2 | >4 | 1 | + | 1.5 | 6 | 20 | + | V |
| 20 | 2 | − | >2 | >4 | 2 | + | 8 | 8 | 17 | + | IV |
| 21 | 2 | − | 2 | >4 | 1 | + | 2 | 6 | 12 | − | V |
| 22 | 2 | − | 1 | >4 | 2 | + | 1.5 | 6 | 20 | + | V |
| 23 | 2 | − | 2 | >4 | 2 | + | 1.5 | 6 | 19 | + | V |
| 24 | 1 | − | 1 | >4 | 2 | + | 2 | 6 | 20 | + | V |
| 25 | 2 | − | 2 | >4 | 2 | + | 2 | 6 | 17 | + | V |
| 26 | 2 | − | 2 | >4 | 2 | + | 2 | 8 | 20 | + | V |
| 27 | 2 | − | 2 | >4 | 2 | + | 1.5 | 6 | 19 | + | V |
By disk diffusion assays following the guidelines and interpretative criteria of the Clinical and Laboratory Standards Institute.2
MIC, minimum inhibitory concentration.
By the gradient diffusion test, 3 strains were susceptible to cefoxitin (MIC=4μg/ml), while the 24 remaining strains were resistant, with MIC values of 6 to 8μg/ml. Moreover, 2 strains were resistant to oxacillin, with MIC values of 4 and 8μg/ml, one strain showed a MIC value=3μg/ml and the remaining ones (n=24) were defined as susceptible with MIC values of ≤2μg/ml (Table 1).
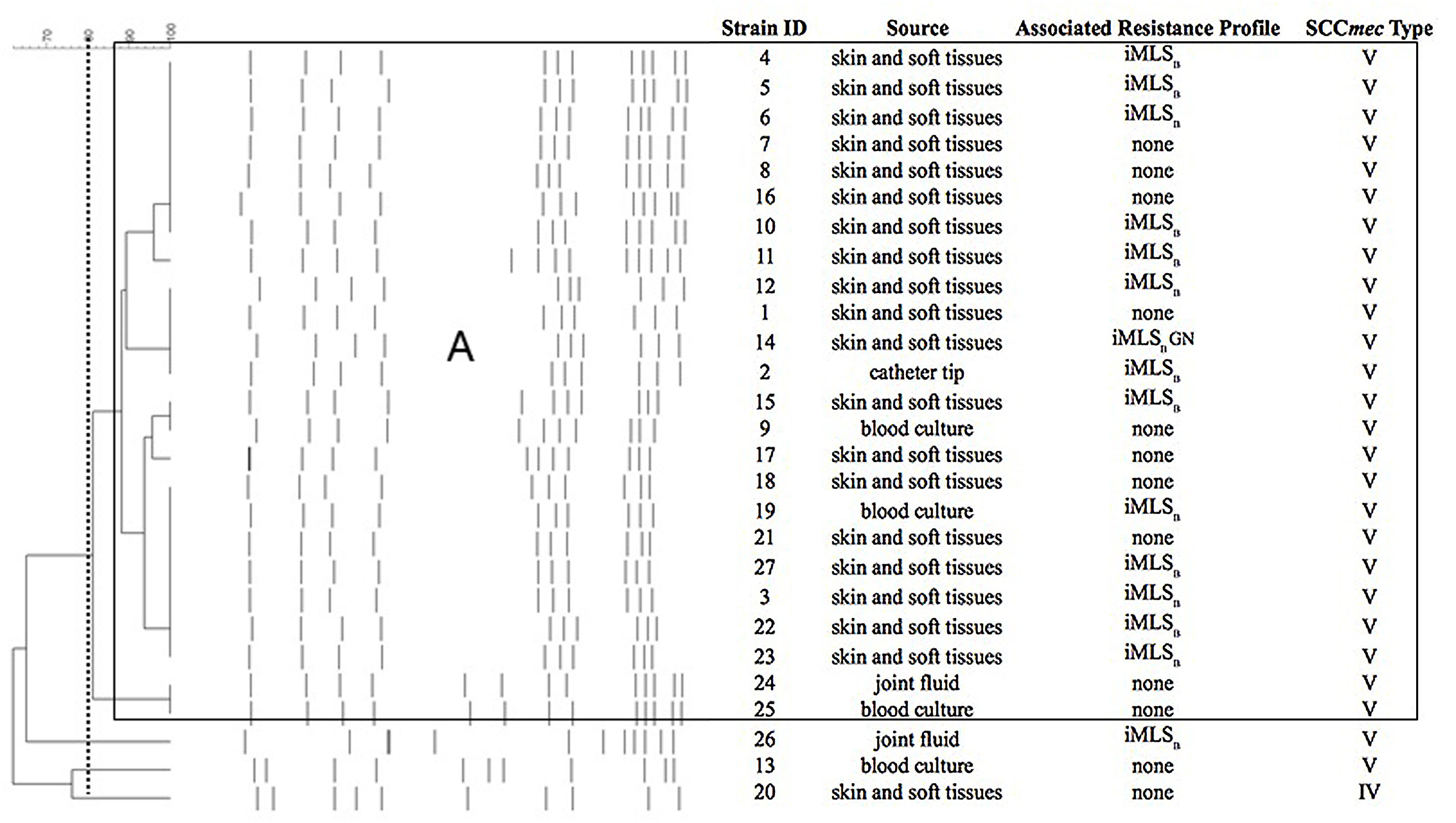
All the strains (n=27) were methicillin-susceptible, 15 displayed the inducible resistance phenotype to macrolides, lincosamides and streptogramin B antibiotics (iMLSB) and one was also resistant to gentamicin (Fig. 1). None of the strains showed resistance to the other antibiotics included in the panels.
Pulsed-field gel electrophoresis (PFGE) patterns and dendrogram of the SmaI-digested genomic DNA obtained from 27 OS-MRSA strains, showing source and associated resistance profile. Band patterns were analyzed by the unweighted pair group method with arithmetic average (UPGMA) using BioNumerics v.7.1.
Dashed line indicates 80%; main box indicates PG A with ≥80% similarity.
iMLSB, inducible resistance to macrolides, lincosamides and type B streptogramin; GN, resistance to gentamicin.
Phoenix® and MicroScan® systems detected resistance to methicillin in 25 and 27 strains, respectively (Table 1).
Twenty-six OS-MRSA strains tested positive for PBP2a.
All strains tested positive for the mecA gene by PCR. Twenty-six OS-MRSA strains carried SCCmec type V and one the SCCmec type IV (see Table 1). PFGE analysis showed that 24 strains belonged to a large cluster or pulsotype (named A, Fig. 1) that carried the SCCmec type V. At the moment and for economic reasons, we performed the MLST assay in only one strain (# 25, isolated from blood culture, SCCmec type V and PT A) (Fig. 1) that corresponded to sequence type 8 (ST8).
This study shows the spread of a set of mecA gene positive S. aureus strains, all misidentified as MSSA by the Vitek 2 system version 6.2. Similar isolates were previously reported from cases of human and animal infections.1,3,10–12,14 In Uruguay, OS-MRSA was first reported by Giudice et al., arising from a discrepancy in the methicillin susceptibility results between two isolates of S. aureus recovered from two clinical samples of the same patient (both strains carrying SCCmec type V and with an identical SmaI-PFGE profile).6 Most strains analyzed in this study (n=24) showed oxacillin MICs of ≤2μg/ml by the gradient diffusion test. These data are consistent with the known fact that oxacillin is not reliable for the detection of methicillin resistance mediated by the mecA gene in S. aureus. Cefoxitin is more reliable but may not yet detect heterogeneous resistance to methicillin. In this sense, three strains were also misidentified as MSSA by the cefoxitin gradient diffusion test and one by the cefoxitin disk assay. Anyway, most strains showed cefoxitin inhibition zones close to the breakpoint value.
As we described before, the discrepancy between these genotypic and phenotypic results could be due, among other reasons, to heterogeneously resistant and inducible MRSA strains.3,4,14,15 We believe that most of the OS-MRSA strains studied here (n=26, all of them PBP 2a-positive) exhibit the oxacillin heteroresistance phenotype.
Only one strain was negative to PBP 2a and could correspond to “stealth methicillin-resistant Staphylococcus” aureus due to mecA gene instability.7 However, to rule out or confirm this hypothesis, it is necessary to carry out at least the complete sequencing of the mecA gene.
Additional problems for the correct identification of this MRSA could be related to the use of automated systems for identification and susceptibility testing. In 2018, the United States Food and Drug Administration recalled Vitek 2 Gram-positive antimicrobial susceptibility testing cards due to false results for some strains of MRSA.5 Taking care of this matter, the local agent of bio Mérieux in Uruguay updated the Vitek 2 software to version 8.01. The 27 strains of OS-MRSA were subsequently retested using this new version, and all of them were correctly identified as methicillin-resistant strains (data not shown).
Unlike the findings of Andrade-Figueiredo et al.1 in which the OS-MRSA strains analyzed were highly diverse by MLST and SmaI-PFGE, the results of PFGE in this work suggest the dissemination of a cluster of S. aureus with SCCmecV in 4 regions of Uruguay. Interestingly, a strain named SA454 with similar phenotypic and genotypic characteristics to most of those described in this work (oxacillin MIC of 1g/ml by the E-test, diameter of the inhibition halo=22mm to cefoxitin disk, mecA and PBP2a positive with SCCmecV, recovered in 2009 from a patient with surgical site infection) was previously reported in Argentina. This suggests that similar or even identical OS-MRSA strains could be present in both countries of the Río de la Plata.3 Other studies that compare in detail the genome of OS-MRSA strains present in both countries are needed to evaluate this hypothesis. One of analyzed 27 strains corresponded to ST8; to better assess the clonality of this set, it would be necessary to carry out this and other procedures of molecular epidemiology on all of them.
We analyzed 27 S. aureus strains carrying the mecA gene that had laboratory problems to be identified as MRSA, probably due to a heteroresistant phenotype. The majority (n=26) exhibited SCCmec type V and belonged to the same pulsotype. The cefoxitin disk and the detection of PBP2a after the induction tests appear to be the most reliable to identify this OS-MRSA. Both procedures are relatively simple and are available in low-complexity microbiology laboratories. However, we must bear in mind that they can yield false negative results; therefore, when there is a suspicion of MRSA for any clinical reason or in strains isolated from severe diseases, it is advisable to use both tests and also detect the presence of the mecA and mecC genes.
Conflicts of interestNone.
FundingThis work was partially supported by a grant of the Comisión Sectorial de Investigación Científica (CSIC), Universidad de la República [initiation to research program – 2017].
Ethical responsibilitiesNone.
We thank Dr. Felipe Schelotto and Jesús Varela for the critical review of the manuscript.