Although Staphylococcus aureus increases its relative abundance in psoriasis when compared with the microbiome of healthy subjects, it is not the most important microorganism underlying this disease. However, there is scant data on the role and molecular features of S. aureus strains in psoriasis; therefore, the aim of this study was to evaluate nasal carriage of this microorganism, its phenotypic and molecular characteristics as well as the impact of host factors on its carriage in psoriatic patients. The presence of S. aureus was analyzed in nasal swabs from 46 healthy volunteers and 50 psoriatic patients by conventional microbiology techniques. Nasal carriage of S. aureus was higher in psoriatic patients than in the control group (37.24% vs 22.98%, respectively), being associated to sex (male), age (adults) and severity of the disease (more frequent in moderate and severe cases). Determination of antibiotic resistance detected 12% of β-lactam resistant isolates, with variable accompanying resistance to macrolides, aminoglycosides and fluoroquinolones. No resistance to rifampicin, vancomycin, mupirocin or trimethoprim/sulfamethoxazole was found.
A preliminary molecular characterization of the isolates was performed by PCR amplification of virulence genes. Molecular characterization of the strains did not reveal a predominant strain in psoriatic patients. Although we established host factors related to increased carriage of S. aureus in psoriatic patients, we could not establish the predominance of one type of strain. Genomic and transcriptomic analysis of the isolated strains would be necessary to address this point.
A pesar de que Staphylococcus aureus incrementa su abundancia relativa en la psoriasis cuando se compara con el microbioma de personas sanas, no es el microorganismo más importante subyacente a la enfermedad. Sin embargo, existen pocos datos sobre el papel y las características moleculares de las cepas de S. aureus en pacientes con psoriasis. Nuestro objetivo fue evaluar la portación nasal de este microorganismo, sus características fenotípicas y moleculares, y el impacto de factores del hospedador sobre dicha portación en estos pacientes. Se analizó la presencia de S. aureus en hisopados nasales de 46 voluntarios sanos y 50 pacientes con psoriasis mediante técnicas microbiológicas convencionales. Se encontró mayor portación en pacientes con psoriasis que en el grupo control (37,24% vs. 22,98%, respectivamente) y esta estuvo asociada al sexo (masculino), la edad (adultos) y la gravedad de la enfermedad (más frecuente en casos moderados a graves). El 12% de los aislamientos de S. aureus mostraron resistencia a betalactámicos, con resistencia acompañante a macrólidos, aminoglucósidos y fluoroquinolonas en grado variable. No se encontró resistencia a rifampicina, vancomicina, mupirocina o trimetroprima/sulfametoxazol.
Se realizó una caracterización molecular preliminar de los aislamientos por amplificación de genes de virulencia mediante PCR. Si bien se identificaron factores relacionados con el hospedador que incrementan la portación nasal de S. aureus en pacientes con psoriasis, la caracterización molecular de las cepas no reveló ninguna característica genotípica predominante asociada a esta afección. Se necesitan más estudios genómicos y transcriptómicos para profundizar en esta caracterización.
Psoriasis (PS) is a chronic inflammatory skin disease that affects 1–3% of the world population irrespective of age or sex, causing a significant impact on the social and working life of psoriatic patients because of the clinical lesions caused by the disease24. Current understanding of its etiology indicates that psoriasis is multi-factorial in origin, with patient genetics, lifestyle and diet contributing to its development and clinical manifestations. From the skin cell biology perspective, an increase in Th7 response has been shown as a key factor in the inflammation process as well as in the proliferation of keratinocytes. For several years the role of microorganisms in the onset and severity of this disease and other chronic inflammatory diseases such as atopic dermatitis (AD) has been discussed. Highly discriminative techniques such as transcriptomic and nucleic acid deep sequencing along with powerful bioinformatics tools are now producing information on the role of the skin microbiome in AD and PS30. A recent report clearly showed differences between the skin microbiome of AD and PS patients, with Staphylococcus aureus being the most predominant bacteria present in the former; however, despite being at increased levels than in healthy volunteers, were less important in the global microbiome imbalance in PS5. However, so far, there is only one study on S. aureus strains specifically present in PS9.
S. aureus frequently colonizes human and animal bodies without signs of overt infection. It is a formidable and versatile pathogen capable of producing various exotoxins such as Panton-Valentine leukocidin (PVL) and enterotoxins, their expression along with that of other virulence traits is positively regulated by agr (accessory regulatory gene system). agr is a global regulator that controls the expression of many virulence factors present in S. aureus. It is known that a functional form of agr is associated with the expression of toxins while the non-functional form is associated with the expression of adhesins. While genes related to colonization are under negative regulation by agr, by switching agr expression on/off, S. aureus changes its lifestyle from a colonizing-prone behavior to a loose, disseminated form31.
The increasing antibiotic resistance (especially against β-lactams) displayed by this pathogen is becoming a major problem in public health. A novel Penicillin Binding Protein (PBP) designated as PBP2A encoded by the mecA gene, was identified as conferring broad β-lactam antibiotic resistance27. This gene is carried on a mobile genetic element (Staphylococcal cassette chromosome mec, SCCmec) containing two discrete regions, mec and ccr (cassette chromosome recombinase) linked by a region designated as J (Junkyard)27. The classification of predominant circulating strains is often based on the analysis of SCCmec, spa type and the presence of genes encoding toxin and virulence factors6.
Although the role of S. aureus as a component of the unbalanced microbiome in AD and psoriasis has been studied, so far, there are no studies on whether a specific S. aureus genotype is predominant in psoriatic patients. As an initial step toward that goal, we herein report the isolation and characterization of S. aureus from a cohort of PS patients and a control group attending a public major hospital in Rosario, Argentina. Although preliminary, this is the first report on this subject in South America and will warrant future studies on the pathogen features responsible for the differences between fast evolving infections and diseases where S. aureus is chronically present.
Materials and methodsStudy cohortA convenience sample of fifty adult patients with plaque psoriasis (25 men and 25 women) treated at Hospital Provincial del Centenario, a public general hospital in Rosario, Argentina, were recruited. Moreover, 46 healthy volunteers (12 men and 34 women) attending the same hospital with no history of altered skin barrier were recruited for this case-control study. Exclusion conditions for the control group were: positive HIV screening test, cancer, or other immunosuppressive conditions, inflammatory skin conditions and antibiotic use within six months prior to sample collection. Patients were assessed via medical visits, interviews, and reviews of medical records by trained dermatologists from the Dermatology Service, Hospital Provincial del Centenario, who collected information on disease severity, comorbidities, and treatments. Samples were taken from October 2016 to October 2017. Personal and demographic data included age and sex. In the case of female volunteers, the use of oral contraceptives was not considered an exclusion factor in spite of reports indicating a possible link between persistent nasal carriage and hormonal contraceptives34. Obesity has been proposed as a factor increasing S. aureus nasal carriage25; however, none of the volunteers in our study was obese. The severity of the disease presentation was assessed based on the scores obtained for BSA (Body Surface Area), PASI (Psoriasis Area and Severity Index) and DLQI (Dermatology Life Quality Index). The definition of moderate-to-severe disease was (PASI, BSA and DLQI>10) and for mild psoriasis (PASI, BSA and DLQI≤10)19. Healthy volunteers did not have a personal history of PS. This study was evaluated and approved by the Bioethics Committee from the Faculty of Biomedical Sciences, Rosario National University.
Specimen collectionDuplicate nasal swab samples were collected using individually packed, sterile cotton swabs. Study participants were asked to refrain from showering and using any substances on their skin (lotion, perfume, make-up, among others) for at least 24h prior to sampling. Swabs were inserted in test tubes containing Stuart medium (Britania, Argentina) and taken to the microbiology lab within 2h of sampling.
Microbiology and drug susceptibility techniquesSamples were spread on Chapman medium (Britania, Argentina) followed by incubation at 37°C for 48h. Possible S. aureus isolates were confirmed by Gram stain and conventional confirmatory techniques (presence of catalase, ability to clog serum). The assessment of resistance to antibiotics was performed by disk diffusion protocols according to CLSI4. The antibiotics tested and concentration used were: cefoxitin (FOX, 30μg), ciprofloxacin (CIP, 5μg), erythromycin (ERY, 15μg), clindamycin (CLI, 2μg), rifampicin (RIF, 5μg), trimethoprim-sulfamethoxazole (TMP/SMX, 25μg), tobramycin (TOB, 10μg), gentamycin (GEN, 10μg), vancomycin (VAN, 30μg) and mupirocin (MUP, 200μg). In the latter two cases, the use of disks was only for the purpose of detecting susceptibility of high level resistance. For isolates that displayed resistance to ERY and susceptibility of intermediate resistance to CLI, testing for induction of inducible clindamycin resistance (ICR) was performed by disk diffusion using the D-zone test.
Molecular characterization of S. aureus isolatesStrain identity and their methicillin resistance was confirmed by multiplex PCR of 16S rRNA, mecA and nuc genes18. Template DNA from each isolate was obtained according to the MLST (MultiLocus Sequencing Typing) protocol as described in https://pubmlst.org/organisms/staphylococcus-aureus/primers. PCR amplification and analysis of chosen genes (see below) were done by standard protocols using the primers listed in Table S1. Determination of agr loci as type I, II, III and IV was performed by multiplex PCR8,9. Detection of ica, lukS/F-PV and sei was carried out by using the primers described in the literature9,16,21. In each case, appropriate positive controls strains (kindly provided by Dr. M. Mollerach, School of Pharmacy and Biochemistry, University of Buenos Aires) were included.
SCCmec typing was performed according to recommendations of the International Working Group on the Classification of the Staphylococcus Cassette Chromosome Elements (IWG-SCC, http://www.sccmec.org). SCCmec molecular typing was assessed by determining mec (A, B, or C) and recombinase (ccr) combinations as described in Refs. 7, 12, 13, 15 (Table S1). Complex ccr1, ccr2 and ccr3 (ccrAB) were determined as described by Ito et al. (Table S1)12. When needed, the presence of Tnp20 in class B mec was tested by using s IS1272 and mecR1 primers (Table S1) followed by amplicon (2kbp) purification26. A diagnostic digestion with two single cutter restriction enzymes (HindIII and XbaI) was chosen to verify the insertion.
Phenotypic assaysFunctionality of agr locus. In order to test the functionality of the agr operon, the evaluation of δ-hemolysin activity was made by cross-streak of the strains under assay with S. aureus RN4220 (a β-hemolysin producer) on Blood Agar Plates33.
Production of biofilm. The ability to form biofilms of the strains under study was measured by a test tube adherence protocol at 37°C on TSB supplemented with dextrose 1% (w/v) and visual inspection at 24, 48 and 72h23. S. epidermidis strain NRS101 and S. aureus strain Newman Δica were included as (+) and (−) controls respectively.
Statistical analysisStatistical analysis was performed using RStudio Version 1.1.463. Odd ratio was determined using epi.2by.2 within the epiR package. Multivariate MCA (Multiple Correspondence Analysis) was obtained by FactoMineR and Factoextra packages (https://cran.r-project.org/web/packages/factoextra/index.html). V and Chi square test were calculated by the catdes function of the FactoMineR package. The adjusted ratio was calculated by the direct method, where the rate expected to be found in the populations under study – provided that all of them had the same composition according to the variable we wanted to adjust –, was calculated. A population designed as “standard”, whose segments corresponded to the factor we wanted to adjust and to which specific rates used for the same segments of the populations under study, were used. Therefore, a number of “expected” cases if the composition of each population was the same for each segment, was obtained. The adjusted rate was calculated as the ratio of expected cases by the total “standard” population20. Group comparison was calculated by the Fisher test using Epi-info (v7.2). Images were edited using Inkscape version 0.92.5.
ResultsAnalysis of S. aureus nasal carriageWe determined the frequency at which S. aureus is carried in the nostrils of psoriatic patients compared to a healthy population. The age and sex distribution of PS and control groups as well as their nasal carriage findings are shown in Table 1. Carriage of S. aureus was statistically more often found in PS patients (20 out 50) than in the control group (8 out 46), with a p value of 0.024, an odd ratio of 3.12. Nasal carriage showed a significant difference when sex was considered, with a higher carriage in male for both groups (60% and 417%, for PS and CG, respectively) and in the different age groups (Table 2). Since age and sex are known variables linked to S. aureus carriage10, we used an adjusted ratio to discard the influence of those two variables; still, S. aureus was more frequently isolated in PS patients (37.24%) than in healthy individuals (22.98%) (Fisher Test, p<0.05).
Staphylococcus aureus carriage by age interval and sex in PS and control groups.
| Group | Age groupsa | Sex | Patient number | Total isolates | Prevalence ratio |
|---|---|---|---|---|---|
| PS | Y | M | 5 | 4 | 4/5 (80%) |
| F | 5 | 2 | 2/5 (40%) | ||
| A | M | 4 | 4 | 4/4 (100%) | |
| F | 5 | 1 | 1/5 (20%) | ||
| S | M | 16 | 7 | 7/16 (43.75%) | |
| F | 15 | 2 | 2/15 (13.33%) | ||
| Total | 50 | 20 | 20/50 (40%) | ||
| C | Y | M | 4 | 1 | 1/4 (25%) |
| F | 10 | 1 | 1/10 (10%) | ||
| A | M | 2 | 1 | 1/2 (50%) | |
| F | 11 | 0 | 0/11 (0%) | ||
| S | M | 6 | 3 | 3/6 (50%) | |
| F | 13 | 2 | 2/13 (15.38%) | ||
| Total | 46 | 8 | 8/46 (17.4%) | ||
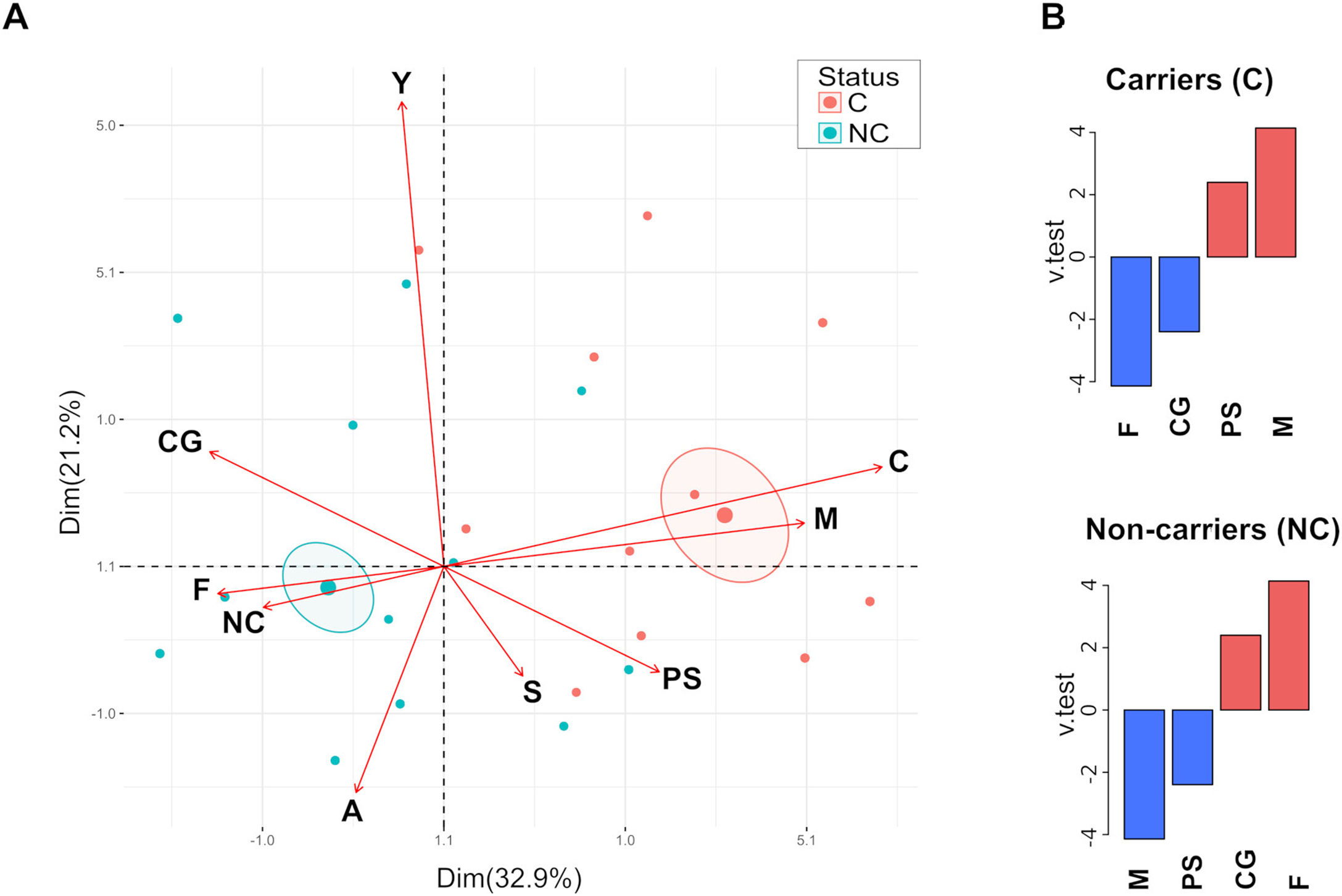
Analysis of the relationship between categorical variables by Multiple Correspondence Analysis (MCA) revealed that carriage is more related to PS and male individuals, whereas the non-carriers are mainly associated with healthy and female individuals (Fig. 1A). Confidence ellipses (p<0.05) around the categories (C and NC) of the variable carriage showed statistically significant differences. Correlation analysis of the variable carriage showed a significant link to the variable sex (p=2.15e−0.5) (p=1.49e−0.2) determined by the Chi square test. Moreover, the analysis of the v test parameter showed that male and PS categories were over-expressed for the carrier group (positive values), whereas female and CG categories were over-expressed for the non-carrier group (Fig. 1B). No correlation was observed for age groups and carriage, neither for PS nor CG groups (Chi square test, p>0.05).
Multiple correspondence analysis. (A) Biplot graphic was generated by using FactoMineR and factoextra in RStudio. Variables analyzed are indicated by arrows; C=carrier, NC=non-carrier, M=male, F=female, Y=young, A=adult, S=senior, PS=psoriasis patients and CG=control group. Confidence ellipses were drawn around the mean of the carriage variable (p<0.05). (B) Variable correlation was analyzed with the catdes function; down and over expressed variables (blue and red bar, respectively) were depicted for carriers and non-carrier groups.
Patients (grouped upon dermatology evaluation using BSA, PASI and DLQI scores) undergoing a moderate to severe form of PS carried S. aureus more frequently than those having a mild form of the disease (58.33% vs 23.08% respectively; Fisher Test, p<0.05) (Table 3). Further analysis showed that male patients with moderate to severe psoriasis showed a high nasal carriage (71.43%) while female patients in the equal severity group showed less nasal carriage (40%). The same analysis for groups suffering a mild form of the disease showed 45.45% and 6.67% nasal carriage respectively (Table 3). The statistical analysis of severity for each sex showed no difference (Fisher exact test. p>0.05).
Relationship between nasal carriage of S. aureus carriage and PS severity.
| Nasal carriage | Sex | Severity | |
|---|---|---|---|
| Mild | Moderate to severe | ||
| NCa | M | 6 | 4 |
| F | 14 | 6 | |
| MSSA | M | 4 | 8 |
| F | 0 | 2 | |
| MRSA | M | 1 | 2 |
| F | 1 | 2 | |
| Total | 26 | 24 | |
We next addressed the profile of antibiotic resistance in both strains isolated from psoriatic patients and from the control group (Fig. 2A). Our findings showed that MRSA was more than twice higher in PS patients (30%) than in the control group (12.5%), followed by resistance to macrolides (35% vs 25% in each group). With regard to macrolide resistance, CLI resistance was always detected by the D-Test and in all cases, was inducible. Only one strain (PS group) was resistant to fluoroquinolones. All the strains were susceptible to RIF, TMP/SMX, MUP and VAN in agreement with our previous studies at the same hospital2. Thus, based on our results, even though there were more MRSA strains in the PS group, the frequency of strains sensitive to all antibiotics is roughly the same (50%) for both groups, indicating that no significant difference in antibiotic resistance was present in both groups. Moreover, the influence of genre in S. aureus nasal carriage has only been observed in MSSA carriers as reported by us and others2,10, whereas this influence has not been observed for MRSA carriage. The prevalence of MRSA in the PS group was 12% for both males and females, whereas for the control group only one MRSA strain was isolated. However, more work is necessary to address this issue (see Discussion).
Schematic representation of antibiotic resistance profiles (A) and virulence genes (B) present in MRSA and MSSA isolated strains. (A) The phenotypic analysis of antibiotic resistance was carried out for MRSA and MSSA strains for each group; R and S represents resistant and susceptible, respectively. (B) The presence or absence of each feature are depicted by (+) and (−), respectively for each group. The agr type, δ-hemolysin activity, the presence of ica, sei and PVL genes and SCCmec type (for MRSA strains) are indicated.
A full genomic typing of the S. aureus strain requires the detection of a large set of genes5 and even genome sequencing which is costly; thus, we identified the SSCmec type, the presence and expression of the agr locus – a global regulator of expression-, and the presence of genes relevant for virulence such as ica, lukS/F-PV and sei as an initial characterization of our isolates.
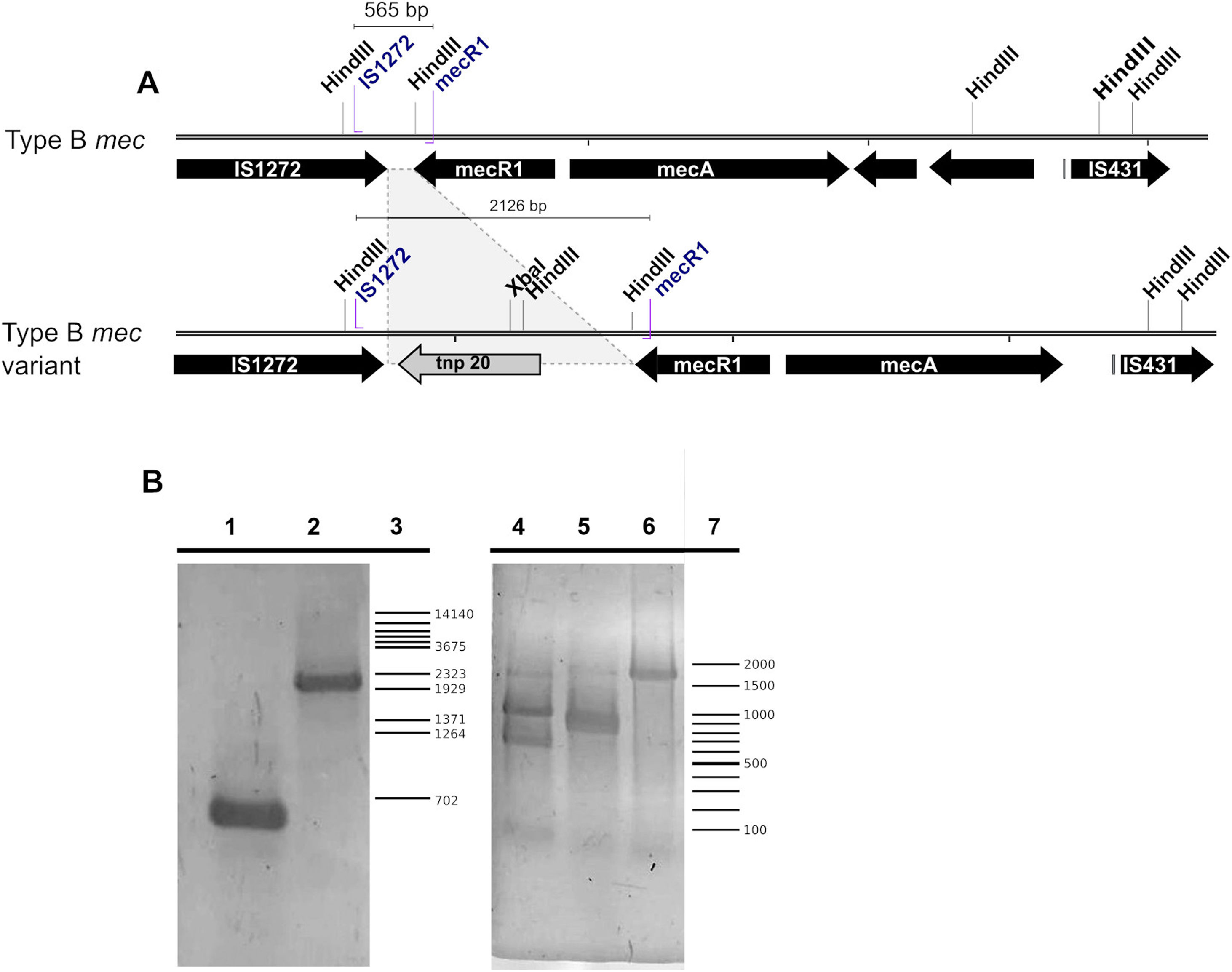
The typing of the SCCmec cassette in each MRSA strain showed that in the PS group five strains contained a recombinase type 2 (A2B2) and a type B mec gene, thus being classified as SCCmec type IV while the remaining isolate contained a recombinase type I (A1B1) and a type B mec, thus being classified as SCCmec type I (Fig. 2B). Of note, one of the five SCCmec type IV strains showed a mecB PCR product of a size larger than the expected (≈2000bp versus 565bp) due to the insertion of a genetic element related to transposase 20 (tnp20) as reported previously26 (Fig. 3A). This was confirmed here by PCR amplification using mecB IS1272 and mecR1 primers, and followed by digestion of the amplicon with HindIII and XbaI, obtaining the expected fragment sizes of 1210, 787, 129 pb and 1115 and 1015 pb, respectively (Table 1) (Fig. 3B). Only one MRSA strain from the control group was identified, which upon PCR analysis showed to contain a type 2 recombinase (A2B2) along with a class B mec, thus being also classified as SCCmec IV.
(A) Scheme of a typical SCCmec type IV class B element and a tnp20 insertion variant. HindIII and XbaI restriction sites and primer hybridization sites are depicted. (B) Agarose gel of the mecB PCR product. Left gel, lane 1, mecB cassette; lane 2, mecB::tnp20 variant; lane 3, λ/BstEII molecular weight marker. The gel on the right shows the amplicon digestion with XbaI (lane 4), HindIII (lane 5) or undigested (lane 6); lane 7, 100bp molecular weight ladder.
Sequence polymorphisms of agr locus yields four types (I–IV); our analysis showed that the agr type I loci is prevalent in the isolates of both PS and control groups (60% and 87.5%, respectively), which agrees with published reports on its frequency28. Meanwhile, agr type II was detected only in three MRSA strain of the PS group, and a combination of two agr locus types I–II and I–III were only identified in strains isolated in the PS group. In most cases, agr was functional as judged by agr dependent δ-hemolysin activity (90% and 75% for PS and CG, respectively). Both the presence of ica and biofilm production was demonstrated in all the strains isolated from both groups. lukS/F-PVL was detected in only a single MRSA strain from each group (Fig. 2B).
Enterotoxins are superantigens that exacerbate the inflammatory response through polyclonal proliferation of T cells1,29. The presence of sei (the most frequently isolated enterotoxin in strains of psoriatics patients9) was detected in both groups although with slightly different frequency (65% and 75%, for PS and CG groups, respectively) (Fig. 2B).
DiscussionThe field of the microbiology of chronic inflammatory skin diseases is developing by the application of deep genomic sequencing, transcriptomics, metabolomics and bioinformatics. A recent example is the first detailed description of microbiome changes in AD and psoriasis as well as of global patterns of cutaneous gene expression in those patients5. While Staphylococcus aureus is the major species present in AD (with the consequent decrease in other beneficial species) and is associated to a specific skin transcriptomic signature of the host, psoriasis presents a weaker association between specific microorganisms and skin gene expression in this disease5. Moreover, comparable results were published by Statnikov et al., who showed that there are increases in the four major skin-associated genera (Corynebacterium, Propionibacterium, Streptococcus and Staphylococcus), both at the lesion sites and at unaffected sites in the counterpart of the body30. This allowed these authors to propose “cutaneotypes” or clusters, of which cutaneotype 2 (Firmicutes–Actinobacteria) was enriched in lesions and healthy skin sites of psoriatic patients30. However, although the weight of S. aureus in the psoriasis microbiome seems to be more diffuse than expected, the use of genomic and transcriptomic tools has overlooked the identity of the “typical” (if any) S. aureus strain in psoriasis. Importantly, there are no reports on WGS for the analysis of S. aureus strains isolated from psoriatic patients. In order to start addressing this point, we analyzed the presence of S. aureus in nasal swabs of psoriatic and healthy patients. Our findings correlate with those of others -reviewed by Totte et al.32 – since we detected higher nasal carriage of this microorganism in psoriatic patients compared to the control group. Multivariate analysis showed that sex has a positive correlation with carriage in psoriatic patients, being males more frequently colonized than females). S. aureus carriage also correlated with the severity of the disease; contrary to what was described by Statnikov et al., who found that no specific taxa could be linked to all the degrees of severity30. Differences in experimental design could explain the differences between the results of both groups.
The profile of antibiotic resistance of S. aureus strains isolated from PS patients and healthy individuals differed in that we found a higher proportion of MRSA carriers in the first group than in the control group (33.3% and 12.5% respectively). This result is in agreement with early reports14. Although neither group showed resistance to VAN, MUP, RIF or TMP/SMX, strains isolated from PS patients were more frequently resistant to macrolides than to fluoroquinolones.
A previous study by this group of S. aureus nasal carriage among health care workers at the same hospital attended by both PS and CG individuals yielded 24% MSSA and 6% MRSA2 while a previous study at a different location in Argentina found 38% MSSA and 4% MRSA22. Other reports in the literature usually studied specific groups by age or occupation (i.e. patients and healthy individuals: medical students, health workers, community residents, young and senior individuals, to name a few) thus preventing direct extrapolation of our results3,17; however, our findings show that the proportion of MRSA strains is clearly higher in patients suffering from psoriasis.
In summary, we have conducted a preliminary phenotypic and molecular characterization of S. aureus in patients suffering from psoriasis. More detailed studies addressing the molecular features (MLST, spa typing, toxin repertoire, resistance to methicillin) of clones present in psoriatic patients would be of value to provide a starting point to identify predominant (if any) circulating clones associated with this skin disease. A recent publication by Gregorio et al.11 describing the whole genome sequencing of 190 CC30-MRSA strains circulating in Argentina provides a very well characterized framework for future work. Furthermore, a transcriptomics approach will be necessary to learn how S. aureus interacts with the host in this chronic skin disease. The strains isolated and reported herein would be very valuable to that end.
Conflict of interestThe authors declare that they have no conflicts of interest
Funding: This work was funding by Agencia Nacional de Promoción de la Investigación, el Desarrollo Tecnológico y la Innovación (Agencia I+D+i), Argentina, through Grant PICT 3795-2017.