Macrolide-resistant Streptococcus pneumoniae emerged in Argentina in 1995, representing 26% of invasive infection isolates in children under 5 years old. The objectives of this study were to describe the prevalence of ermB and mefA genes in macrolide-resistant S. pneumoniae isolates from acute otitis media (AOM) and to determine their genetic relatedness. Between May 2009 and August 2010, 126 S. pneumoniae isolates from 324 otherwise healthy children with a first episode of AOM were included. Twenty six of these isolates (20.6%) were resistant to erythromycin. Most frequent serotypes were: 14 (46.2%), 6A (23.1%), 19F (7.7%) and 9V (7.7%). Twenty (76.9%) carried the mefA gene, 5 (19.2%) have the ermB gene, and 1 (3.9%) both ermB + mefA. Ten clonal types were identified, mostly related to Sweden15A-25/ST782 (SLV63), CloneB6A/ST473 and England14-9/ ST9. This is the first study assessing the mechanisms of macrolide resistance in pneumococci isolates from pediatric AOM in Argentina and their genetic relatedness.
Streptococcus pneumoniae resistente a macrólidos emergió en la Argentina en 1995 y representa el 26 % de los aislamientos de infecciones invasivas en niños menores de 5 años. El objetivo de este trabajo fue describir la prevalencia de ermB y mefA en neumococos resistentes a macrólidos aislados de niños con otitis media aguda (OMA) y determinar su relación genética. En 15 meses se incluyeron 126 neumococos aislados de 324 niños previamente sanos, con un primer episodio de OMA. Veintiseis (20,6 %) eran resistentes a eritromicina. Los serotipos más frecuentes fueron los siguientes: 14 (46,2 %), 6A (23,1 %), 19F (7,7 %) y 9V (7,7 %). Veinte eran portadores del gen mefA, 5 del gen ermB y el restante portaba ambos genes. Se identificaron 10 tipos clonales, la mayoría relacionados con los clones Sweden15A-25/ST782 (SLV63), CloneB6A/ST473 y England14-9/ST9. Este es el primer estudio en determinar los mecanismos de resistencia a macrólidos en neumococos aislados de OMA en la Argentina y en describrir su relación genética.
Acute otitis media (AOM) is by far the most common disease caused by Streptococcus pneumoniae and one of the most frequent diagnoses in children under 2 years old. One out of every three children has at least one episode of AOM during the first three years of life1.
Over the last years, resistance to penicillin in S. pneumoniae has increased in Latin America and worldwide10,11. Consequently, macrolides were used as initial empiric therapy for community-acquired respiratory tract infections11. Unfortunately, resistance to macrolides in S. pneumoniae has also evolved rapidly and in many countries the rate of S. pneumoniae resistant to macrolides exceeded the level of resistance to penicillin2,8. Results from a global international surveillance project (PROTEKT, 1993–2003) showed an increase in macrolide resistance from 31% in 1999 to 36.3% in 200311. Macrolide resistance in S. pneumoniae causing infectious diseases in pediatric patients emerged in Argentina in 1995 and firstly increased to 1.4% between 1995–1997 and later to 6% in the period 1998–20016. Nowadays, according to the Argentinian SIREVA II surveillance data, macrolide resistance reached 30.2% in 201110. Four different mechanisms of resistance to macrolides have been described: antibiotic efflux, mediated by mefA gene; target-site modification by methylation of 23S rRNA due to erm genes; mutations in riboproteins L4 or L22 and mutations in domain II or V of 23S rRNA. The mefA gene, displaying an M-phenotype, has been associated with resistance to 14- and 15-membered macrolides. The ermB gene is known to confer cross-resistance to macrolides, lincosamides and streptogramins B, yielding the MLSB- phenotype9.
The objectives of the present study were to determine the frequency of macrolide-resistant S. pneumoniae isolates from AOM, the prevalence of ermB (ribosomal methylase) and mefA (efflux pump) genes and to determine their genetic relatedness.
Between May 2009 and August 2010, 324 immunocompetent children under 120 months (median age: 8 months) with a first episode of AOM with purulent exudates in the middle ear were included in this study. Immunocompromised patients and those with chronic otitis media were excluded. All middle ear specimens obtained through tympanocentesis were cultured under standard conditions7. One hundred and thirty three pneumococci were isolated from 324 children suffering from AOM (41%), but 126 were available for this study.
Pneumococcal strains were serotyped by the Neufeld Quellung reaction using antiserum from the Statens Serum Institut, Copenhagen, Denmark.
Minimal inhibitory concentrations (MIC) were determined by the agar dilution method using Mueller Hinton agar (Difco, BD, USA) supplemented with 5% sheep blood and incubated overnight at 35°C. MIC values of penicillin, amoxicillin, cefotaxime, erythromycin and clindamycin were interpreted according to Clinical and Laboratory Standards Institute recommendations. Susceptibility to chloramphenicol, tetracycline, ofloxacin, vancomycin and trimethoprim-sulfamethoxazole was tested by the disk diffusion method according to the CLSI guidelines. Phenotypic characterization of macrolide resistance was performed by the double-disk diffusion method (D test), using disks of erythromycin (15μg) and clindamycin (2μg) to discriminate isolates expressing the M- and MLSB- phenotype (inducible or constitutive). MICs were interpreted according to the CLSI guidelines5.
PCR assays for detection of mefA and ermB genes and pulse-field gel electrophoresis (PFGE) were performed as previously described (6). PFGE patterns were categorized using the Tenover criteria14 and compared to international clones (http://www.sph.emory.edu/PMEN/). Multilocus sequence typing (MLST) was carried out as previously described, with five selected strains of each dominant PFGE pattern and those associated with international clones (www.mlst.net/).
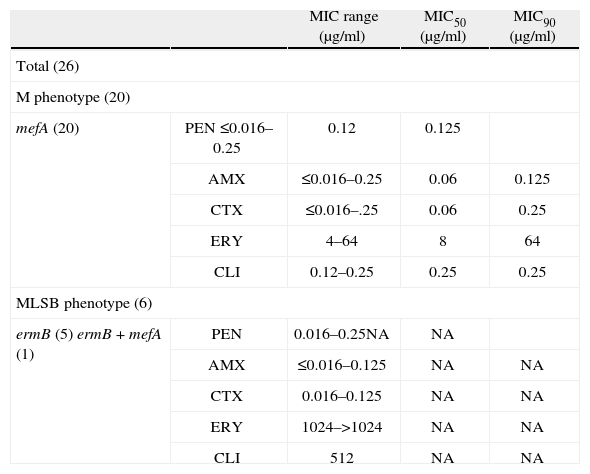
Twenty six of the 126 available pneumococci (20.6%) were classified as erythromycin-resistant and were further studied. Although macrolide resistance in our country has been increasing steadily since 199510, these resistance levels do not exceed erythromycin resistance from invasive infections in other countries4. Most frequent serotypes among macrolide-resistant pneumococci from AOM were (n; %): 14 (12; 46.3) and 6A (6; 23.1); followed by 19F (2; 7.7), 9V (2; 7.7), 6B (1; 3.8), 19A (1; 3.8), 33F (1; 3.8) and nontypeable (1; 3.8) compared with 14 and 6B, found in a previous study of invasive infections6. Almost in coincidence with our results, in Bedouin and Jewish children with AOM, predominant serotypes with macrolide resistance were: 6A, 6B, 14, 19A and 19F13 and in Bulgarian children, the predominant serotypes were: 6B, 19A and 19F12. MIC range, MIC50 and MIC90 for penicillin, amoxicillin, cefotaxime, erythromycin and clindamycin of the 26 isolates are shown in table 1. In 65.4% of cases, erythromycin resistance was associated with penicillin MICs between 0.12 and 1μg/ml. Breakpoints for penicillin were those recommended for oral penicillin V5. We have to emphasize that applying the current breakpoints of CLSI, all isolates were susceptible to amoxicillin, which is the antibiotic recommended for the empirical treatment of AOM in our country. All isolates were susceptible to cefotaxime (MIC ≤ 1μg/ml), and also to chloramphenicol, ofloxacin and vancomycin when tested by the disk diffusion method, using the breakpoints recommended by the CLSI5.
Minimal inhibitory concentration and correlation among phenotypes and genotypes of macrolide-resistant Streptococcus pneumoniae.
| MIC range (μg/ml) | MIC50 (μg/ml) | MIC90 (μg/ml) | ||
| Total (26) | ||||
| M phenotype (20) | ||||
| mefA (20) | PEN ≤0.016–0.25 | 0.12 | 0.125 | |
| AMX | ≤0.016–0.25 | 0.06 | 0.125 | |
| CTX | ≤0.016–.25 | 0.06 | 0.25 | |
| ERY | 4–64 | 8 | 64 | |
| CLI | 0.12–0.25 | 0.25 | 0.25 | |
| MLSB phenotype (6) | ||||
| ermB (5) ermB + mefA (1) | PEN | 0.016–0.25NA | NA | |
| AMX | ≤0.016–0.125 | NA | NA | |
| CTX | 0.016–0.125 | NA | NA | |
| ERY | 1024–>1024 | NA | NA | |
| CLI | 512 | NA | NA | |
PEN, penicillin; AMX, amoxicillin; CTX, cefotaxime; ERY, erythromycin; CLI, clindamycin; NA, not applicable.
Resistance to erythromycin was recorded in 26 (20.6%) isolates of S. pneumoniae from AOM, mainly due to the presence of the mefA gene. Resistance to tetracycline and trimethoprim-sulfamethoxazole was observed in 13 (50%) and in 15 (57.7%) macrolide-resistant isolates, respectively.
Twenty of the 26 macrolide resistant isolates (76.9%) expressed the M-phenotype and were positive for the mefA gene, while 6 (23.1%) expressed the constitutive MLSB- phenotype. Five of them were positive for the ermB gene and one was positive for both genes (table 1). As has been previously described, isolates displaying the MLSB-phenotype showed higher MICs of erythromycin and clindamycin than those with the M-phenotype. All S. pneumoniae serotype 14 isolates harbored the mefA gene (11/12 expressed the M-phenotype and 1/12 expressed the MLSB-phenotype due to the presence of both mefA and ermB genes). All serotype 6A isolates harbored the mefA gene and expressed the M-phenotype. Our results are consistent with previous Argentinian data of macrolide-resistant S. pneumoniae isolated between 1993 and 20016. However, in European countries, macrolide-resistant S. pneumoniae strains are predominantly ermB positive12 and in South Korea dissemination of macrolide-resistant S. pneumoniae was due to isolates containing the ermB gene alone or concomitantly with the mefA gene15. None of the macrolideresistant isolates was found to be mefA and ermB negative. Other macrolide resistance mechanisms, such as mutations in the 23S rRNA or alterations in ribosomal proteins L4 and L22 have not been investigated.
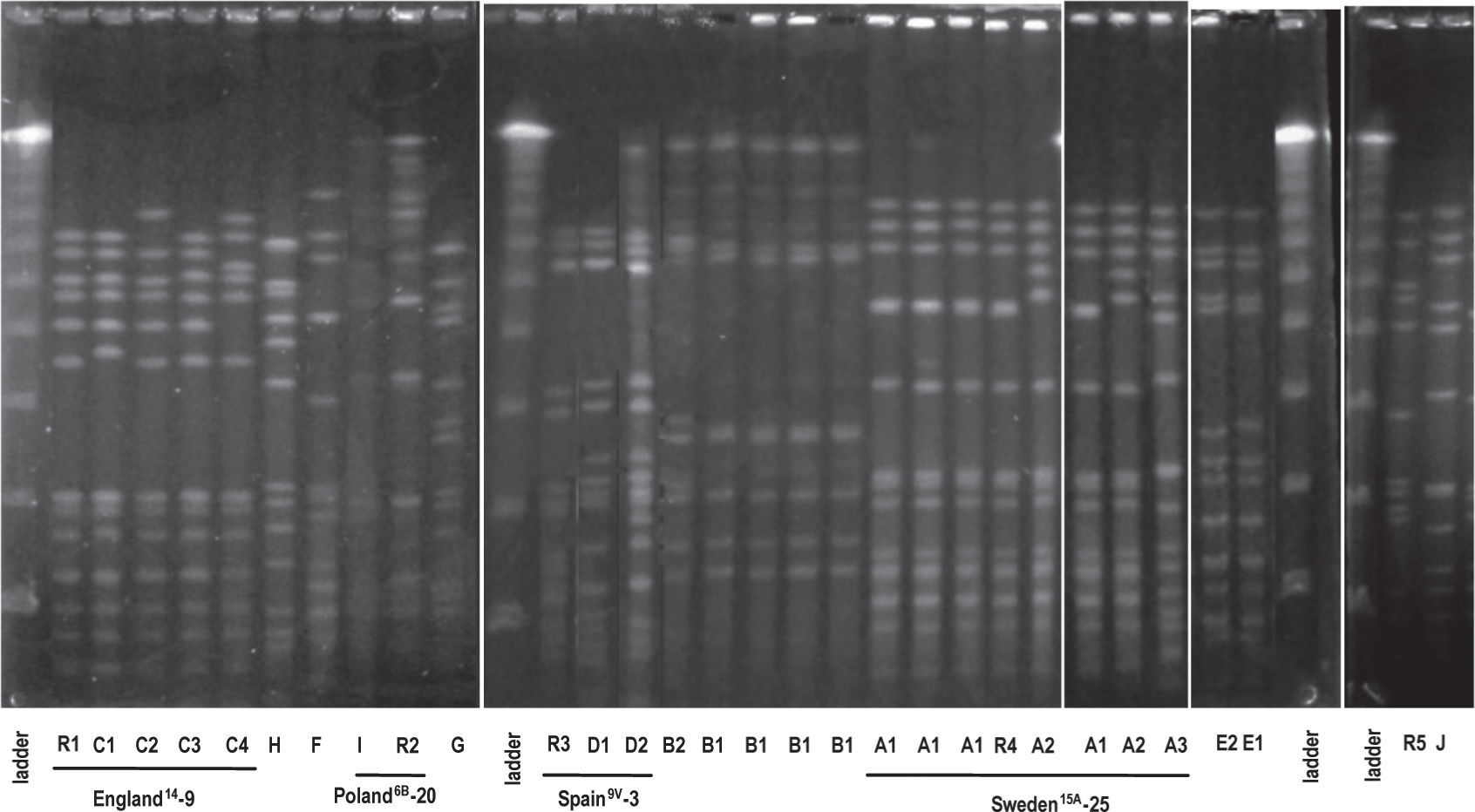
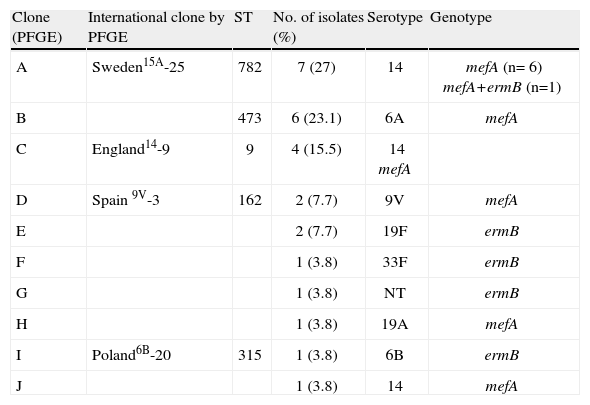
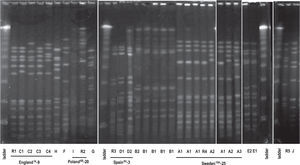
Ten (10) clonal types (A to J) were defined by the SmaIPFGE pattern. (fig.1). Seventeen isolates were grouped into three clones: clone A n:7, clone B n:6, and clone C n:4. The PFGE pattern of clones A and C were related to Sweden15A-25 and England14-9 international clones defined in PMEN while clone B6A was not related to any international clone. The remaining nine isolates were grouped into seven clones, of which only two were related to international clones: Spain9V-3 and Poland6B-20 (table 2). In the study of Corso et al. referring to invasive infections, the emergency of erythromycin-resistant S. pneumoniae was principally due to the England14-9, Poland6B-20 and Spain9V-3 international clones 6, while in the present study both the Sweden15A-25 and CloneB6A clones were the major ones (table 2). This could be the consequence of differences in spreading trends of clonal types producing AOM with respect to those producing invasive infections.
Comparison of macrolide-resistant isolates with international clones.
| Clone (PFGE) | International clone by PFGE | ST | No. of isolates (%) | Serotype | Genotype |
| A | Sweden15A-25 | 782 | 7 (27) | 14 | mefA (n= 6) mefA+ermB (n=1) |
| B | 473 | 6 (23.1) | 6A | mefA | |
| C | England14-9 | 9 | 4 (15.5) | 14 mefA | |
| D | Spain 9V-3 | 162 | 2 (7.7) | 9V | mefA |
| E | 2 (7.7) | 19F | ermB | ||
| F | 1 (3.8) | 33F | ermB | ||
| G | 1 (3.8) | NT | ermB | ||
| H | 1 (3.8) | 19A | mefA | ||
| I | Poland6B-20 | 315 | 1 (3.8) | 6B | ermB |
| J | 1 (3.8) | 14 | mefA |
NT, non-typeable.
MLST was performed on clones which showed higher frequencies and on those that were associated with some international clones. All the isolates belonging to the Sweden15A-25 clone were ST782 (SLV63) and clustered to CC63. CloneB6A was ST473 and clone England14-9 was ST9. Clone Spain9V-3 was ST162, a single locus variant of ST156. Clone Poland6B-20 was ST315 (table 2). Although PFGE patterns were identical to the Sweden15A-25 and Spain9V-3 clones, when we performed MLST, we noted that our isolates had different ST but belonged to highly related clonal complexes.
Sweden15A-25 (CC63) is a worldwide disseminated clone usually showing capsular switching (serotype 19F or 19A). This clone has been described in Spain as a cause of meningitis and among multidrug- and ciprofloxacin-resistant strains with different serotypes (14, 19A, 19F, and 23F). In previous European studies4, the Sweden15A-25 clone was the most frequently found clone among erythromycin-resistant pneumococci (www.mlst.net). The MLST database includes several ST63 isolates recovered from AOM in France. However, this database does not contain ST782 isolates from AOM. In the present study, the Sweden15A-25 clone was also the most frequently found clone expressing serotype 14, probably due to a previous capsular switching event.
Alonso et al. studied pneumococcal isolates causing AOM in a region of Northern Spain between 1999 and 2010. The authors detected that clones ST63 and ST156 were only occasionally present in their region, the last one associated with ERY-susceptible isolates1. Bowers et al. reported the dominance of multidrug-resistant CC271 clones in macrolide-resistant pneumococci in Arizona, principally due to the dual genotype (mefE/ermB)3. Among the mef positive isolates, they found two belonging to the ST156 clone, one of them showing serotype 9V, similar to clone D isolates found in our study, and one isolate belonging to the ST162 clone. Among the ermB positive isolates, they found eight ST63 and two ST315, with serotype 6B, similar to clone I.
This study was carried out before the introduction of the PCV13 vaccine in our immunization schedule in 2012; therefore, it deserves to be repeated later, to evaluate the vaccine effectiveness and the possible spread of new S. pneumoniae clones in AOM.
Although macrolides or lincosamides are not first-line antibiotics for the treatment of AOM, the knowledge about the prevalence of different macrolide-resistant mechanisms allowed us to assess and eventually predict their dissemination among pneumococci.
Although in our country we have data on the evolution of macrolide-resistant S. pneumoniae since 1995 from invasive infections and AOM 6, this is the first study assessing the mechanisms of macrolide resistance and genetic relatedness of S. pneumoniae isolated from pediatric patients suffering from AOM in Argentina.
Ethical approvalThis research was approved by the Ethics Committee and the Dirección Asociada de Docencia e Investigación (Teaching and Research Associated Board) at the Hospital de Pediatría “Prof. Dr. Juan P. Garrahan”.
FundingThe research leading to these results has received funding from the Seventh Framework Programme of the European Community under Grant Agreement No. HEALTHF3- 2009-223111.
Conflicts of interestThe authors declare that they have no conflicts of interest.