We evaluated the microbial composition of water kefir grains and beverage over the course of one year to determine whether the number and type of microorganisms changed over the time. Bacteria and yeast colonies with different morphologies were isolated from water kefir and their antimicrobial activity was evaluated against Paenibacillus larvae and Ascosphaera apis. A chemical characterization of kefir was also carried out. Our results confirmed that bacteria and yeasts were more numerous in kefir grains compared with those in the beverage. The counts of microorganisms declined, although an important microbial community was still present in kefir after the long storage period. Eleven strains which inhibited bee pathogens were isolated from kefir. Genotypic results demonstrated that these isolates included Lentilactobacillus hilgardii, Lentilactobacillus buchneri and Saccharomyces cerevisiae. Thus, water kefir may be an innovative source of potential probiotic strains for bee nutrition in order to control honeybee diseases.
Evaluamos la composición microbiana del kéfir de agua durante un año para determinar si la cantidad y el tipo de microorganismos cambiaban con el tiempo. Se aislaron colonias de bacterias y de levaduras con diferentes morfologías, y su actividad antimicrobiana se evaluó frente a Paenibacillus larvae y Ascosphaera apis. También se realizó una caracterización química del kéfir. Nuestros resultados confirmaron que las bacterias y las levaduras eran más numerosas en los gránulos de kéfir en comparación con la parte líquida. Los recuentos de microorganismos disminuyeron, aunque una cantidad igualmente importante se encontró en el kéfir después de un año. Se aislaron del kéfir once cepas que inhibieron los mencionados patógenos de abejas. Los resultados genotípicos demostraron que estos aislamientos eran Lentilactobacillus hilgardii, Lentilactobacillus buchneri y Saccharomyces cerevisiae. Por lo tanto, el kéfir de agua podría ser una fuente innovadora de potenciales cepas probióticas para contribuir a la nutrición y sanidad de las abejas.
Honeybee health can be affected by inadequate nutrition; moreover, nutritionally stressed colonies are more vulnerable to parasites and show higher mortality rates. Antibiotics and pesticides that beekeepers apply in colonies could generate resistant pathogens and cause disturbances in the balance of the beneficial bee microbiota. Thus, probiotics represent a biologically sustainable strategy to control honeybee diseases and also improve the health of bee colonies2. In this sense, water kefir, a fermented beverage, may be an innovative source of potential probiotic microorganisms to control honeybee diseases.
In previous studies, we evaluated whether the beverage obtained from water kefir could be used as a supplement to feed honeybees and to enhance their health6,9,13. We reported the effect of water kefir on the survival, weight, and size of bee larvae. We also analyzed whether larvae could develop into the stages of pupa and imago13. Based on those studies, we hypothesized that water kefir could be an innovative source of potential probiotic strains for bee nutrition. Therefore, the aim of this study was to evaluate, over the course of one year, the microbial composition of both, water kefir grains and the beverage, in order to determine whether the number and type of microorganisms remain stable over time. A chemical characterization of water kefir was also performed. Several bacteria and yeast isolates were obtained from water kefir so as to prepare a collection of probiotic strains intended for bee nutrition.
Water kefir was prepared with 60g of grains which were inoculated into 400ml of a sugar solution (14g/l of unrefined sugar) with two raisins and a slice of lemon. The beverage was contained in a glass jar with a non-hermetic cover. The fermentation process was conducted at room temperature (23±2°C) for 48h10. After fermentation, the grains were washed with distilled water before the next culture incubation. Samples of water kefir were stored at room temperature (23±2°C). Three replicates of water kefir were prepared for the assay.
Water kefir was chemically analyzed after three days of fermentation according to Laureys and De Vuyst10 with few modifications. Briefly, acidity (AOAC, 1995)1, pH (measured using a digital pH meter Hanna Instruments HI98103), ethanol (percentage of alcohol content by volume (% ABV) determined using a densitometer (Macotest, Argentina)), mineral content (sodium, potassium and magnesium assessed by inductively coupled plasma optical emission spectrometry (ICP-OES)), total soluble solids (measured using a handheld laboratory refractometer (Atago N1 (Brix 0-32% Japan)), carbohydrates (Antrona method), proteins (Kjeldahl method), and dry matter content (AOAC, 1995)1 were determined in the beverage while ash and dry matter content were measured in kefir grains. All chemical tests were performed in triplicate.
Kefir grains and beverage were sampled eight times during the course of one year (2019–2020) in order to evaluate their microbial composition. Ten millilitres of beverage and 10g of grains were homogenized into 90ml of sodium citrate 2% (initial suspension). Decimal serial dilutions of each sample were performed using sterile distilled water. The viable microorganisms count was carried out by the spread plate technique. The diluted suspensions were plated on Petri dishes with de Man, Rogosa, and Sharpe agar medium (MRS, Bioclar, Argentina) to enumerate presumptive lactic acid bacteria (LAB). Plates were incubated at 37°C under microaerophilic and anaerobic conditions for five days. Yeasts were enumerated on fungi and yeast agar medium (HyL, Britania, Argentina). Plates were incubated at 25°C in an aerobic atmosphere for three days10. After the incubation period, colony-forming units (CFU) of LAB and yeasts were counted. Results were expressed as log CFU per ml of beverage or per gram of kefir grains.
Presumptive LAB and yeast colonies with different morphologies were selected from the plates used for the viable count, in order to prepare a collection of probiotic strains intended for bee nutrition. These isolates were subjected to phenotypic characterization to describe them as LABs or yeasts, according to the criteria reported by Audisio et al.2. Based on the results obtained, the isolates were evaluated against Paenibacillus larvae (UB-CIDEFI PL105, AR1, AR2, LF1) and Ascosphaera apis (181, 186, 188, 191) which belonged to the CEMIBA-UNLP collection.
Each isolate was screened against A. apis using a combination of the central disk test assay12 and the agar-well diffusion method4. Briefly, circles of mycelium of A. apis were obtained from colonies growing on plates of YGPSA medium and were placed in the center of new plates with wells containing the isolates. Plates were incubated at 28°C in aerobic conditions. The agar-well diffusion method was performed for P. larvae. Plates of MYPGP medium were incubated at 35°C under aerobic conditions. After 4 and 7 days, plates were checked for any growth inhibition of A. apis or P. larvae. Inhibition activity was recorded in a four-level scale as follows: (0mm) no inhibition of the growth of A. apis or P. larvae; (5–7mm) medium inhibition; (7–9mm) good inhibition; and (9–12mm) very good inhibition of the growth of the pathogen strain.
Isolates showing the greatest antagonism against both bee pathogens were identified by molecular analysis. Genomic DNA of the bacterial isolates and PCR reactions were done according to Syrokou et al.14. Universal primers P0 (5′-GAGAGTTTGATCCTGGCTCAG-3′) and P6 (5′-CTACGGCTACCTTGTTACGA-3′) were used to amplify a fragment of about 1500bp of the 16S rRNA gene. Amplified PCR products were sequenced bidirectionally using the commercial service of Macrogen, Korea (http://macrogen.com/). The obtained sequences were used to query the SeqMatch tool of the Ribosomal Database Project II5.
Yeast genomic DNA extraction and PCR were performed according to Avin et al.3. The nucleotide sequences of partial 18S, complete internal transcribed spacer 1 (ITS1), 58S, internal transcribed spacer 2 (ITS2), and partial 28S were amplified by PCR using primers ITS1F (5′-CTTGGTCATTTAGAGGAAGTAA-3′) and ITS4 (5′-TCCTCCGCTTATTGATATGC-3′). The products were analyzed as previously described for bacteria.
Analysis of variance (one-way ANOVA) was run to identify differences between microbial enumerations in the beverage and grains over time. Means were tested by the Fisher's least significant difference (LSD) test (α=0.05). Student t-test was performed to compare the mean counts (log CFU) of bacteria and yeasts between beverage and kefir grains, as well as the initial and final number of microorganisms after one year. An α value <0.05 was considered statistically significant. All statistical analyses were done using InfoStat Version 20137 and Epidata Version 4.2 – 20168.
The results of the chemical analyses done in water kefir revealed that the beverage samples showed a titratable acidity of 0.777g/100±0.005ml and a pH value of 3.44±0.05. Total carbohydrate content was 1.806±0.05mg/ml, total protein content was 0.017% and dry matter content was 0.51±0.04% w/v. Ethanol concentration increased from 0.20% ABV after 24h, to 1.19% ABV after 72h of fermentation. Total soluble solids ranged from 3.4 to 1.73 °Brix from the initial time to 72h of fermentation. Mineral content of water kefir was 77.6mg/l sodium, 165.4mg/l potassium, and 21.1mg/l magnesium. Grain samples exhibited a mean dry weight of 17.12±1.24% w/w and the ash content was 0.69±0.01% (g ash/100g dry grains).
Chemical properties of water kefir, like microbiological characteristics, depend on factors such as manufacturing process, incubation, and storage conditions. This is presumably the reason why, some results obtained from our kefir samples differed from those of other studies. The protein levels of the studied samples were lower than those determined by some authors, and the carbohydrate and ash content were different from values previously reported11.
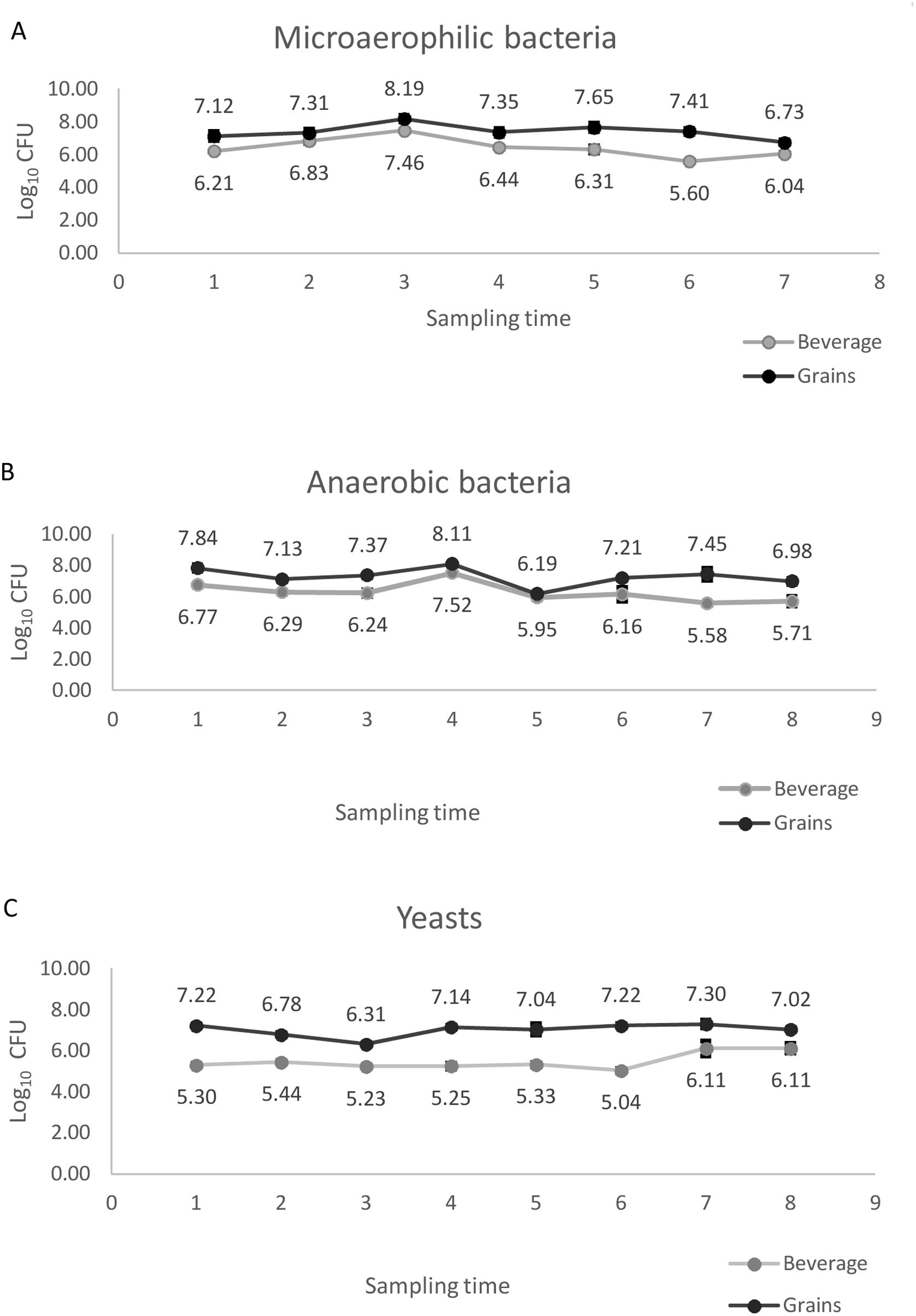
The average viable counts of bacteria and yeasts isolated from water kefir grains and the beverage during the course of a year were registered. The number of bacteria and yeasts isolated from kefir grains was contrasted with the number obtained from the beverage. The count of each group of microorganisms was significantly higher (p<0.01) in kefir grains compared with that of the beverage. The average viable counts of microaerophilic bacteria, anaerobic bacteria, and yeasts were 7.39±0.07 log CFU/g, 7.28±0.14 log CFU/g, and 7.00±0.11 log CFU/g of water kefir grains, respectively, and 6.41±0.08 log CFU/ml, 6.27±0.15 log CFU/ml, and 5.48±0.14 log CFU/ml of kefir beverage, respectively.
The count of each group of microorganisms isolated from grains and the beverage was evaluated during a year to determine whether the number of CFU remained stable or changed (Fig. 1). The one-way ANOVA test revealed marked differences between the beverage and kefir grains in the populations of microorganisms analyzed during the storage time. The number of microaerophilic and anaerobic bacteria in the beverage was maximum after five months of storage and then tended to decrease. In contrast, the count of yeasts remained constant until the final period when it increased. On the other hand, the count of bacteria in kefir grains tended to decrease over the whole conservation period, showing a significant decrease after five months. The yeast population was stable during the first 5 months of storage, considerably increasing afterwards.
The initial number of microorganisms was compared with the final one in order to establish if it remained stable over one year of storage. In the kefir grains, the count of microaerophilic bacteria, anaerobic bacteria, and yeasts declined significantly (p<0.01) after 12 months. In the beverage, the number of microaerophilic and anaerobic bacteria showed no statistical differences (p≥0.05) after one year, whereas the count of yeasts increased significantly (p<0.01) after the storage period.
Bacteria and yeast colonies with distinct morphologies were selected, obtaining a total of 54 isolates from water kefir. Based on the phenotypic characterization, 30 presumptive LAB isolates were recognized as Gram positive, catalase-negative, oxidase-negative, non-motile, non-nitrate reducer, and glucose-fermentative without CO2 production, and lactose-fermentative strains. Twenty-four yeast isolates were recognized as catalase-positive, oxidase-positive, non-nitrate reducer, and non-lactose fermentative strains.
Bacteria and yeasts preselected according to the phenotypic characterization were evaluated against P. larvae and A. apis. With regard to A. apis, of the 20 anaerobic bacteria isolates (presumptive LAB), 9 produced no inhibition (45%); 8 produced medium inhibition (40%); and 3 produced good inhibition (15%). Of the 24 yeast isolates, 6 produced no inhibition (25%) and 18 produced medium inhibition (75%). Regarding P. larvae, of the 20 anaerobic bacterial isolates, 17 produced no inhibition (85%), and 3 produced medium inhibition (15%). Of the 24 yeast isolates, 7 produced medium inhibition (29.2%), 12 produced good inhibition (50%); and 5 produced very good inhibition (20.8). Out of 10 microaerophilic bacteria, none of them inhibited any A. apis or P. larvae strains.
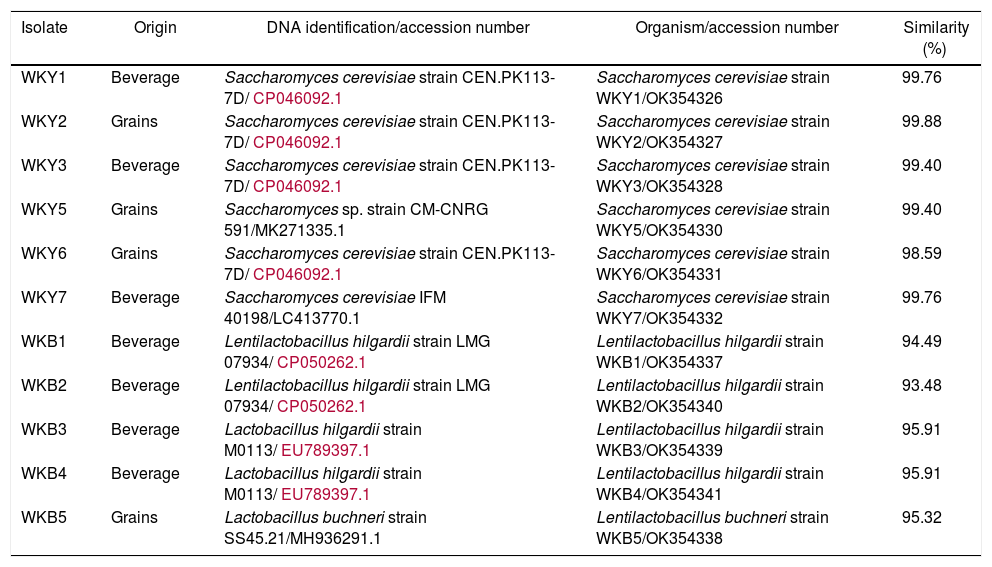
Eleven isolates showing antagonism against P. larvae and A. apis were identified by molecular analysis (Table 1). The comparative analysis of the 16S rRNA sequences revealed that the presumptive LAB isolates are closely related to Lentilactobacillus sp. (formerly Lactobacillus): Lentilactobacillus hilgardii and Lentilactobacillus buchneri (formerly Lactobacillus hilgardii and Lactobacillus buchneri)15. The presence of these species has been previously reported. Lentilactobacillus hilgardii is commonly found in water kefir10, whereas L. buchneri has been less frequently reported in water kefir. On the other hand, the nucleotide sequences of partial 18S, ITS1, 58S, ITS2, and partial 28S have revealed that the yeast isolates are closely linked to Saccharomyces cerevisiae. This is the most prevalent yeast species in water kefir14.
Molecular identification of anaerobic bacteria and yeast strains isolated from water kefir.
| Isolate | Origin | DNA identification/accession number | Organism/accession number | Similarity (%) |
|---|---|---|---|---|
| WKY1 | Beverage | Saccharomyces cerevisiae strain CEN.PK113-7D/CP046092.1 | Saccharomyces cerevisiae strain WKY1/OK354326 | 99.76 |
| WKY2 | Grains | Saccharomyces cerevisiae strain CEN.PK113-7D/CP046092.1 | Saccharomyces cerevisiae strain WKY2/OK354327 | 99.88 |
| WKY3 | Beverage | Saccharomyces cerevisiae strain CEN.PK113-7D/CP046092.1 | Saccharomyces cerevisiae strain WKY3/OK354328 | 99.40 |
| WKY5 | Grains | Saccharomyces sp. strain CM-CNRG 591/MK271335.1 | Saccharomyces cerevisiae strain WKY5/OK354330 | 99.40 |
| WKY6 | Grains | Saccharomyces cerevisiae strain CEN.PK113-7D/CP046092.1 | Saccharomyces cerevisiae strain WKY6/OK354331 | 98.59 |
| WKY7 | Beverage | Saccharomyces cerevisiae IFM 40198/LC413770.1 | Saccharomyces cerevisiae strain WKY7/OK354332 | 99.76 |
| WKB1 | Beverage | Lentilactobacillus hilgardii strain LMG 07934/CP050262.1 | Lentilactobacillus hilgardii strain WKB1/OK354337 | 94.49 |
| WKB2 | Beverage | Lentilactobacillus hilgardii strain LMG 07934/CP050262.1 | Lentilactobacillus hilgardii strain WKB2/OK354340 | 93.48 |
| WKB3 | Beverage | Lactobacillus hilgardii strain M0113/EU789397.1 | Lentilactobacillus hilgardii strain WKB3/OK354339 | 95.91 |
| WKB4 | Beverage | Lactobacillus hilgardii strain M0113/EU789397.1 | Lentilactobacillus hilgardii strain WKB4/OK354341 | 95.91 |
| WKB5 | Grains | Lactobacillus buchneri strain SS45.21/MH936291.1 | Lentilactobacillus buchneri strain WKB5/OK354338 | 95.32 |
In summary, after one year of storage, microorganisms remained in kefir, and the highest number of these microorganisms were detected in kefir grains. Bacterial isolates were identified as Lentilactobacillus hilgardii and L. buchneri, and yeasts were identified as Saccharomyces cerevisiae. Our results demonstrated that these isolates inhibited A. apis and P. larvae. Thus, water kefir may be an innovative source of potential probiotic strains for bee nutrition to control honeybee diseases. Forthcoming assays will include massive genotype sequencing techniques to describe the complex microbiome community of water kefir.
Conflicts of interestThe authors declare that they have no conflicts of interest.