Infection of water buffaloes (Bubalus bubalis) with bovine viral diarrhea viruses (BVDV) has been confirmed in several studies by serological and molecular techniques. In order to determine the presence of persistently infected animals and circulating species and subtypes of BVDV we conducted this study on a buffalo herd, whose habitat was shared with bovine cattle (Bossp.). Our serological results showed a high level of positivity for BVDV-1 and BVDV-2 within the buffalo herd. The molecular analyses of blood samples in serologically negative animals revealed the presence of viral nucleic acid, confirming the existence of persistent infection in the buffaloes. Cloning and sequencing of the 5′ UTR of some of these samples revealed the presence of naturally mix-infected buffaloes with at least two different subtypes (1a and 1b), and also with both BVDV species (BVDV-1 and BVDV-2).
La infección de los búfalos de agua (Bubalus bubalis) con los virus de la diarrea viral bovina (BVDV) ha sido confirmada mediante técnicas serológicas y moleculares en trabajos anteriores. Con el fin de determinar la presencia de animales persistentemente infectados y las especies y subtipos circulantes de BVDV en esta especie animal se realizó un estudio sobre una manada de búfalos de producción mixta con ganado bovino (Bossp.). Nuestros resultados serológicos mostraron un alto nivel de positividad frente a BVDV-1 y BVDV-2 dentro de la manada de búfalos. El análisis molecular sobre muestras de sangre de los animales serológicamente negativos reveló la presencia de ácido nucleico viral, lo que confirma la existencia de búfalos persistentemente infectados. El clonado y la secuenciación de la región 5 ‘UTR de algunas de las muestras obtenidas de búfalo reveló la presencia de coinfección natural con al menos dos subtipos diferentes de BVDV (1a y 1b) y con las especies virales BVDV-1 y BVDV-2.
Pestivirus is a genus of viruses of the Flaviviridae family that comprises four well-defined species, bovine viral diarrhea virus 1 (BVDV-1), bovine viral diarrhea virus 2 (BVDV-2), border disease virus (BDV) and classical swine fever virus (CSFV). Although the natural hosts of pestiviruses are cows, sheep and pigs, they are also known to cross species barriers infecting a wide range of animals in the order Artiodactyla8.
BVDV has worldwide distribution and causes various clinical syndromes in cattle including diarrhea, mucosal disease, and reproduction dysfunctions (abortion, teratogenesis, embryo resorption, fetal mummification and stillbirth). However, the acquisition of an infection (before 150 days of gestation) by a pregnant cow can lead to the birth of an immunotolerant calf with a persistent infection (PI)3.
Persistent BVDV infection in ruminant species, other than bovines, has been previously reported5,11. In a previous study, we have also reported the presence of viral antigen and nucleic acid in different organs of water buffaloes suggesting persistently infected (PI) status6. With respect to this previous result, the present study was carried out to study the presence of PI animals and circulating subtypes of BVDV in a herd of water buffaloes (Bubalus bubalis) that belongs to a farm with a mixed production system (buffaloes and cattle). This farm is located in the northeastern region of Argentina (San Martin Department, Corrientes Province), and both species share the same field and have never been vaccinated against BVDV.
We sampled the serum and blood of 38 of 80 buffaloes and 10 cows, twice, with a 6-month interval between both sample periods, to confirm the occurrence of infection in this animal population. All sampled buffaloes were female aged around 2 years old.
We performed a virus neutralization test against BVDV to screen the presence of serologically positive and negative animals. This assay was carried out for either BVDV-1 using a NADL strain, or BVDV-2 using the local cytopathic isolate 61380 (AF417986) separately, as described in the OIE Manual of Standards for Diagnostic Tests and Vaccines.
The virus neutralization tests showed a high level of serological positivity for BVDV in the analyzed population. While buffaloes showed 84.2% (32/38) of positivity, both for BVDV-1 and BVDV-2, 100% of the cow samples (10/10) were positive for these two viral species. This high serological prevalence observed in buffaloes was similar to that reported by others1, and in this particular case, it could be due to the close contact between buffaloes and other bovines in the examined farm.
After a 6-month period, we took a second serum sample from the same previously sampled buffaloes. The analyses of these samples showed no differences in neutralizing serological values. Only 6 of the 38 buffaloes (# 2, 17, 24, 26, 27, and 32) remained negative for one or both viral species throughout both sampling periods. This result is suggestive of a possible PI status of these animals - the maintenance of seronegative status within a 6-month period in animals in a herd with circulating BVDV is not expected. Therefore, in order to confirm these results, we isolated the viral nucleic acid from blood samples of these seronegative buffaloes using TRIzol Reagent (Invitrogen) following the manufacturer's instructions. After a reverse transcription (RT) step, we performed a PCR on the 5′ UTR, using the previously reported pan-pestivirus primer pair 324/32612.
All PCR amplicons showed a band of the expected size of 288bp. After a purification step using the Qiagen Gel Purification Kit™, the amplicons were cloned in a pGEM-T vector system™ (Promega). Some amplicons were not successfully recovered after the purification step (# 26, 27 and 32).
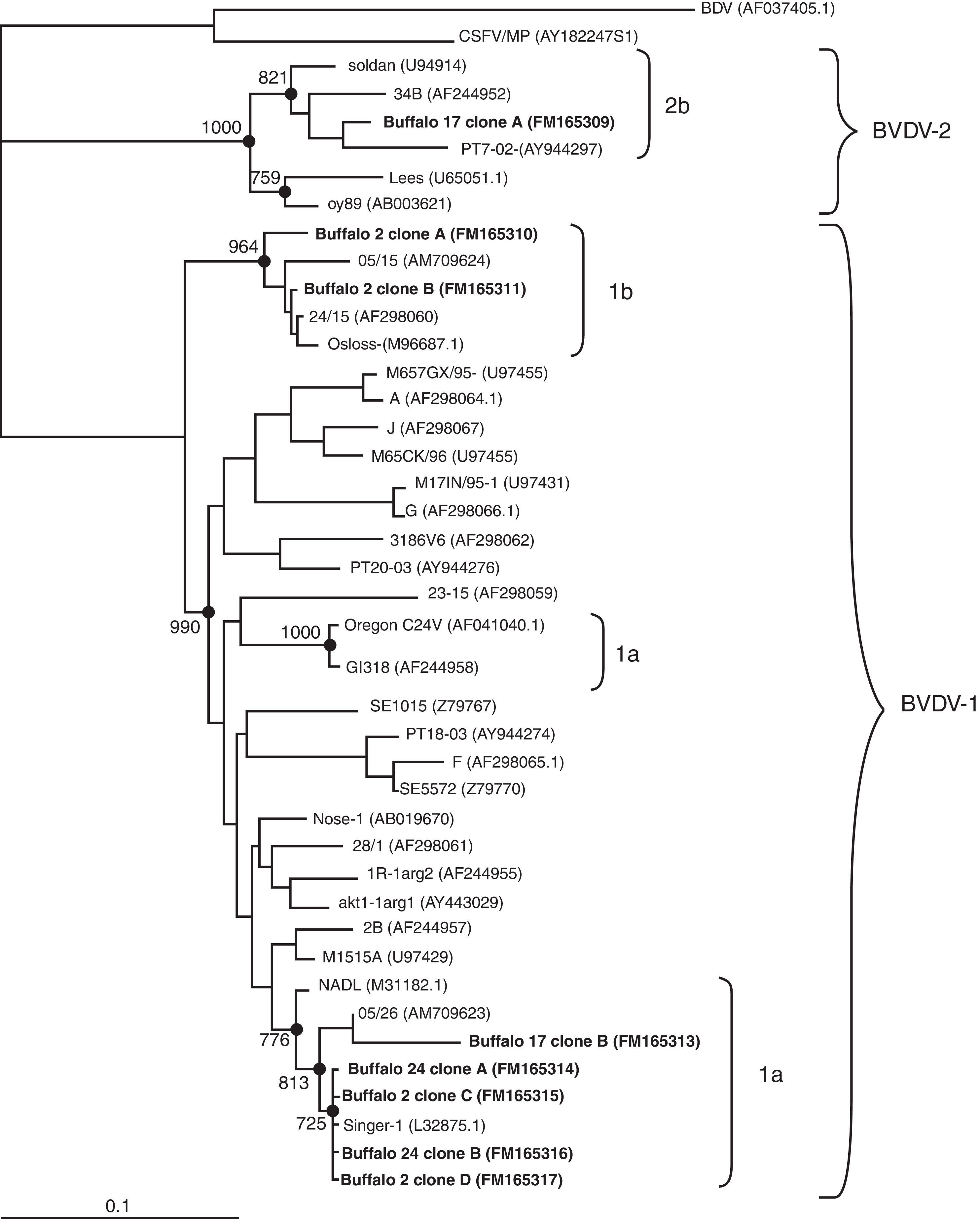
We transformed Escherichia coli DH5α competent cells with the recombinant plasmids and then we selected 10 different clones from each sample to be sequenced in an automated ABI PRISM 377 DNA sequencer. All different sequences obtained from the clones were compared against representative pestivirus strains deposited in the GenBank. The sequence analysis of the clones of two buffaloes clustered within BVDV-1 (buffaloes numbered 2 and 24). However, while all the clones from buffalo 24 belonged to subtype 1a, 50% of the clones from buffalo 2 clustered within 1a and the other 50% within 1b (Fig. 1). This result suggests that a naturally mix-infection was present in this animal. While clones of subtype 1a, belonging to buffaloes 2 and 24, grouped together with the Singer strain, clones of subtype 1b were closely related to isolate 05/15 (AM709624), previously detected and reported in buffaloes6. In addition, the molecular analyses of the clones from buffalo 17 showed the presence of both BVDV-1 and BVDV-2. The analysis of these sequences revealed they are closely related to isolate 05/26 (AM709623), which was previously obtained from a BDVD-infected buffalo organ and other sequences belonging to subtype 2b (Fig. 1)6.
Phylogenetic tree constructed by the Neighbor Joining method showing the relationships among BVDV based on 5′UTR region sequences. The tree was constructed with the sequences of representative clones of each sample. Phylogenetic relationships were analyzed using the Kimura 2-p. Nodes with significant bootstrap values (>70%) over 1000 replicates are indicated. The bar represents the relative genetic distance. Genotype groups are indicated by brackets on the right hand side. Accession numbers are shown between parentheses. BVDV detected in Argentinean buffalo are shown in bold.
Summarizing, we amplified BVDV from blood samples of six buffaloes that remained seronegative to the virus, in a herd with evidence of virus circulation during a 6-month period. In two of three of these animals, we detected, by clone sequencing, the presence of one or two species and/or subtypes of BVDV in the same animal. To our knowledge and for the first time this unexpected result confirms the existence of a natural mixed infection of different viral species and/or subtypes, in water buffaloes. Moreover, this ratio of two of three buffaloes with a mixed infection suggests that the level of mixed infection in a buffalo herd could be frequent. Although the occurrence of infection with both subtypes of pestiviruses (BVDV 1a or 1b) in other ruminants other than bovines was previously described4,10, natural co-infection with different species or subtypes of BVDV was not reported in any animal species before. In recent years, some authors provided evidence that experimental infection of cows between days 60 and 90 of gestation with multiple BVDV isolates concomitantly, resulted in the birth of six PI calves where two of them harbored and shed more than one challenge virus2.
Although cloning of PCR amplicons is usually used in molecular studies of viral population, it has not been commonly used to analyze the viral population in studies of natural infections with BVDV. This methodology allowed us to detect the circulation of different variants of BVDV strains in some of these animals. During the 6-month period we observed that buffalo 24 and buffalo 17 remained seronegative to BVDV 1 and BVDV 1 and 2, respectively. The permanence of the seronegative status in addition to a high level of serological prevalence within the herd is an evidence that supports the presence of PI animals9. These results are consistent with the BVDV detected in each blood sample of buffalo 24 (FM165314, FM165316) and buffalo 17 (FM165313, FM165309). Moreover, we detected both BVDV 1a and 1b (FM165310, FM165311, FM165315 and FM165317) circulating in buffalo 2 who was seropositive for BVDV-1 during both sampling periods. We cannot dismiss, however, that one of the two viral species or subtypes sampled after the 6-month period was due to a recently acquired infection in a previously infected animal.
In summary, the present study reports the existence of natural coinfection with different subtypes and species of BVDV. This issue could be a source for the generation of natural recombinants. The existence of recombination in BVDV and other flavivirus has been previously reported supporting the hypothesis that natural co-infection is not a rare event7. The evidence presented in this study highlights the importance of the buffalo as a possible viral reservoir, mainly in countries where a mixed production is practised and governments design national vaccination programs to control the disease. Further studies on these matters will lead to a better understanding of the epidemiology of the virus, and will collaborate to focus on the control program strategy.
Ethical disclosuresProtection of human and animal subjectsThe authors declare that no experiments were performed on humans or animals for this study.
Confidentiality of dataThe authors declare that no patient data appear in this article.
Right to privacy and informed consentThe authors declare that no patient data appear in this article.
Conflict of interestThe authors declare that they have no conflicts of interest.
The authors thank Dr. Mónica Jacobsen, Betiana Parody and Valentina Ferretti for their revision and correction of this manuscript. We also thank the Sequencing Service of the IMYZA Institute for technical assistance. This work was fully supported by BID1201/OC-AR 08-09627.