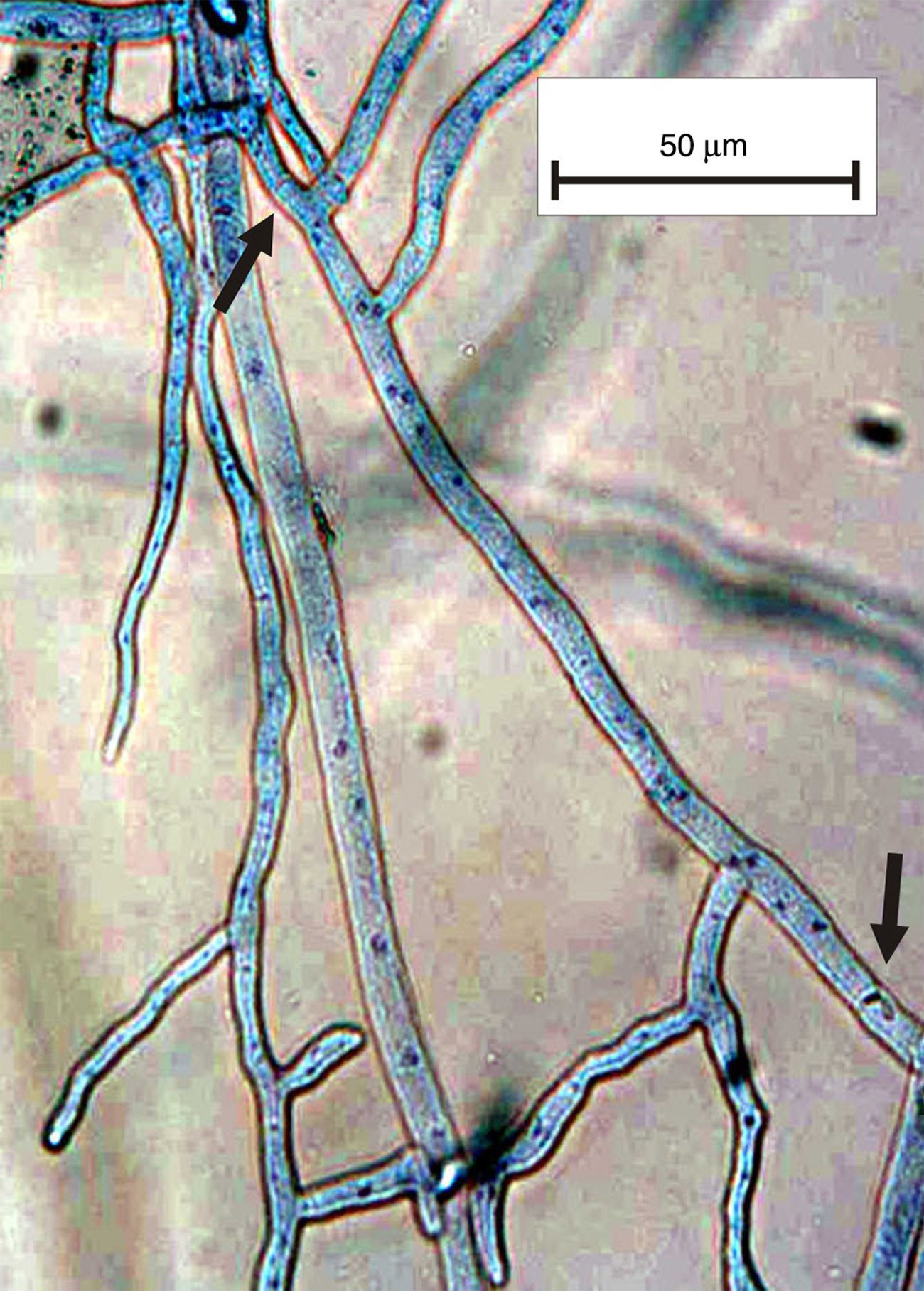
The genus Rhizoctonia DC (1805) has long been studied as an important soilborne pathogen that causes a wide variety of symptoms because it is a non-specialized pathogen3. Rhizoctonia sensu lato is characterized by the lack of conidiogenous cells and this taxon is composed of two groups based on the number of nuclei per cell: the multinucleate group that belongs to Rhizoctonia s. str. and the binucleate group that belongs to Ceratorhiza5. Currently, other authors consider the group a Ceratobasidium–Rhizoctonia complex7 and divide it into two groups: BNR (binucleate Rhizoctonia-like) and MNR (multinucleate Rhizoctonia-like)9. Many methods are used to observe the number of nuclei in fungal cells, e.g. safranine O, aniline blue, HCl-Giemsa. Some of these methods apply a staining solution involving laborious, time-consuming procedures that require no equipment (Fig. 1). Other methods use fluorophores, which are rapid and precise1,2,4,6,8,10.
Since a reliable and rapid method was needed to explore the MNR and BNR groups associated with rice crops in Argentina, we applied an accurate technique to observe the number of nuclei in the strain cells belonging to the Ceratobasidium–Rhizoctonia complex.
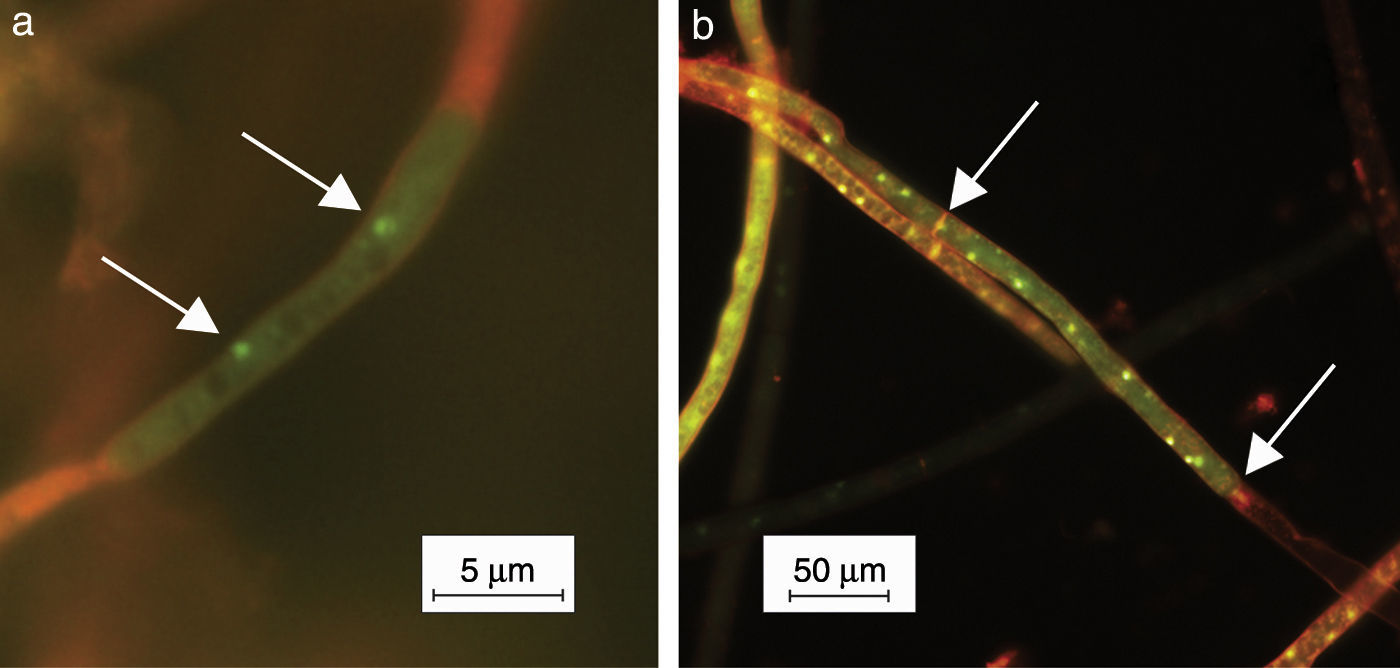
Small portions of mycelia grown on PDA for 24h were submerged in 0.01% acridine orange (Sigma–Aldrich, USA) aqueous solution during 10s. The method applied was a modification of the Yamamoto and Uchida's staining method11, the latter consisting of veronal buffer. The treated mycelium was observed under epifluorescence light using an OLYMPUS BX 51 microscope (Olympus, Japan). Digital photographs were taken using the Cool Snap-Pro System (Media Cybernetics, USA) (Fig. 2). The nuclei showed green fluorescence and the other cell components orange fluorescence. Thirty isolates from a collection of 36 were multinucleate and 6 were binucleate.
In comparison with the other methods mentioned above, the present method is characterized by easy handling and lower cost.
To our knowledge, this is the first time this methodology has been applied to observe nuclei in cells of Rhizoctonia isolates obtained from rice cultivars from Corrientes province, Argentina.
Ethical disclosuresProtection of human and animal subjectsThe authors declare that no experiments were performed on humans or animals for this study.
Confidentiality of dataThe authors declare that no patient data appear in this article.
Right to privacy and informed consentThe authors declare that no patient data appear in this article.
Funding, Financial support and acknowledgementsThe present research was supported by the Governments of Argentina (Instituto Nacional de Tecnología Agropecuaria) and Japan (Japan International Cooperation Agency) in a bilateral collaboration program.
Conflict of interestThe authors declare that they have no conflicts of interest.