Cildáñez stream (in Matanza-Riachuelo basin, Buenos Aires) is one of the most polluted watercourses of Argentina, containing a mixed contamination from agricultural and industrial wastes. The application of water bioremediation processes for this kind of effluent will require microorganisms with a high tolerance to contamination. In this sense, obtaining higher contaminant-resistant microalgae lines is widely desired. In this study, adaptive laboratory evolution (ALE) and random mutagenesis were used to obtain Chlorella vulgaris LMPA-40 strains adapted to grow in polluted water from the Cildáñez stream. The ALE process was performed by 22 successive subcultures under selective pressure (Cildáñez wastewater alone or with the addition of phenol or H2O2) while random mutagenesis was performed with UV-C radiation at 275nm. Not all the cell lines obtained after ALE could adapt enough to overcome the stress caused by the Cildáñez wastewater, indicating that the process is quite random and depends on the stressor used. The best results were obtained for the Cildáñez wastewater adapted cells (Cild 3 strain) that were more resistant than the original strain. The concentration of protein, Chlorophyll A, Chlorophyll B, and carotenoids in the Cild 3 ALE evolved strain was higher than that of the control strain. However, this strain exhibited half of the lipid content compared to the same control strain. Interestingly, these alterations and the acquired tolerance may be reversed over time during storage. These findings suggest that the acquisition of novel cell lines could not be permanent, a fact that must be considered for future trials.
El arroyo Cildáñez (en la cuenca Matanza-Riachuelo) es uno de los cursos de agua más contaminados en Argentina. Este arroyo presenta contaminación mixta proveniente de residuos agrícolas e industriales. En este sentido, existe un amplio interés en la obtención de líneas de microalga más resistentes a la contaminación para usar en procesos de biorremediación. En este trabajo, se empleó la evolución adaptativa de laboratorio (ALE) y la mutagénesis aleatoria para obtener nuevas variantes de la cepa de Chlorella vulgaris LMPA-40 adaptadas para crecer en agua contaminada del arroyo Cildáñez. El proceso ALE se realizó mediante 22 subcultivos sucesivos bajo presión selectiva (agua contaminada del arroyo Cildáñez sola o con el agregado de fenol o H2O2), mientras que la mutagénesis aleatoria se realizó con radiación UV-C a 275nm. No todas las líneas celulares obtenidas mediante ALE pudieron adaptarse lo suficiente para superar el estrés provocado por el agua contaminada del Cildáñez, lo que indica que el proceso es bastante aleatorio y depende del estresor utilizado. Los mejores resultados se obtuvieron con las células adaptadas al agua contaminada del Cildáñez (cepa Cild 3) que fueron más resistentes que la cepa original. La concentración de proteínas, clorofila A, clorofila B y carotenoides en la cepa evolucionada Cild 3 fue mayor que en la cepa control. Sin embargo, esta cepa Cild 3 exhibió la mitad del contenido de lípidos en comparación con la misma cepa control. Curiosamente, estas alteraciones y la tolerancia adquirida pueden revertirse con el tiempo durante el almacenamiento. Estos hallazgos sugieren que la adquisición de nuevas líneas celulares no podría ser permanente y este hecho debe tenerse en cuenta en ensayos futuros.
Water is becoming a limiting resource aggravated by the increase in population growth, pollution, and climate changes. The reduction in the availability of drinking water makes it essential to study new methodologies to optimize its management. Chlorella vulgaris is a fast-growing photosynthetic microalga that can be cultured in several aquatic environments12 including contaminated ones. This plasticity of growth allows this microalga to be used for decontaminating water such as that of the Cildáñez stream. This stream belongs to the Matanza-Riachuelo basin (MRB), one of the most densely populated and industrialized regions of Argentina. The Cildáñez stream presents serious urban environmental problems related to the dumping of domestic and industrial effluents, and the lack of land use planning, making this a wastewater reservoir7.
Microalgae can remove pollutants such as nitrogen forms, phosphorus, microorganisms13, as well as carcinogenic chemicals like phenol from polluted wastewater11,23,35. Moreover, their high performance in utilizing sunlight, CO2, and water to produce biomass allows their reuse, in line with a growing circular economy. Despite the various advantages that microalgae culture offers, there are still several challenges to overcome to scale up the production process in an economically viable way, among them, greater resistance thresholds for organic pollutants such as phenol, and inorganic contaminants such as hydrogen peroxide, should be mentioned.
Adaptive laboratory evolution (ALE) is a laboratory process based on the evolution of microorganisms followed by the selection of individuals with effective mutations and/or recombination that can adapt or survive under certain environmental conditions (for a review, see30). It consists in exposing cultures to stress conditions for long periods (from weeks to years), which results in the accumulation of DNA mutations, leading to the rapid evolution of a strain to gain desirable characteristics2,3,16,22,31. This technique can be used for improving the adaptation of organisms to contaminated environments.
On the other hand, random mutagenesis consists in exposing an organism to a mutagenic agent, which can be chemical, physical (such as UV-C irradiation), or biological. This process generates random DNA changes, some of which may entail characteristics of interest. Only by screening the mutants obtained, improved strains may be identified and selected. A recent review on random mutagenesis techniques applied to microalgal cell factories, with a particular focus on physical and chemical mutagens, mutagenesis conditions, and mutant characteristics can be found in Bleisch et al.4.
The aim of this work was to obtain C. vulgaris cell lines with enhanced resistance to grow in contaminated water samples from the Cildáñez stream through ALE alone, with the addition or not of two stressors (phenol and hydrogen peroxide) or combined with random mutagenesis.
Materials and methodsMaterialsThe native Argentinian strain LMPA-40 of C. vulgaris (Biological Data National System, SNDB-173) used and the cell lines obtained were maintained in a mixotrophic culture in MS medium supplemented with sucrose (3%) and 1mg/l indole acetic acid (IAA), at pH 5.6/5.7, with weekly subcultures to fresh medium (MS)7. Cultures were incubated at 24±2°C with PAR lighting (400μmol photon/m2/s) with a 16-h photoperiod in an orbital shaker at 100rpm.
The water samples from the Cildáñez stream in the Matanza-Riachuelo basin (ACCM) were collected by the Environmental Protection Agency of the Government of the Autonomous City of Buenos Aires (APRA) during the spring of 2020.
ALE experimentsALE experiments were carried out in Erlenmeyer flasks (150ml) containing 30ml of Cildáñez stream water. The initial inoculum was prepared by resuspending C. vulgaris cells collected from 10ml-saturated cultures in 20ml of the treatment solutions, which were:
- -
Cildáñez stream water (Cild) (no stressor added)
- -
Cild+phenol (Phen, 160mg/l)
- -
Cild+H2O2 (H2O2, 100g/l)
The ALE process lasted 110 days, 22 successive subcultures under selective pressure were performed during this period (every 5 days).
Phen and H2O2 concentrations were selected based on the results obtained previously (data not shown).
ALE plus random mutagenesisA fourth treatment to induce random mutagenesis was performed with UV-C radiation (I). This treatment began 50 days after the initiation of the ALE assay in Cildáñez samples. Irradiation was repeated at 5-day intervals, coinciding with the moment of subculturing (12 irradiation sessions in a period of 60 days). Irradiation was performed in Petri dishes with UV-C radiation (OSRAM PURITEC® UV-C Low Pressure Lamp HNS™ L 36W 2G11, 275nm) for 12.5min from 30cm, based on the results obtained previously (data not shown).
Isolation and purification of the surviving C. vulgaris cell lines at the end of the ALE processAt the end of all processes (irradiation and chemical stressors), 1ml aliquots of the final cultures were serially diluted (1:10 to 1:1000) and used to isolate cell lines of green algae. Briefly, MS media supplemented with ampicillin (10mg/l) and streptomycin (10mg/l) was used for isolation in three rounds of culturing, combining subsequent solid agar media (where 200μl of culture was spread using a Drigalski spatula) and liquid cultures (started from green colonies picked up from the plate). The cultures were incubated for 72h at 22±2°C with a 16-h photoperiod. After this period, three green colonies were selected from each treatment based on morphological characteristics under microscopic observation, and named Cild 1, Cild 2, Cild 3, Phen 1, Phen 2, Phen 3, H2O2 1, H2O2 2, H2O2 3, I 1, I 2, I 3. These cell lines were maintained in MS medium for 3 months before carrying out the growth kinetics tests.
Characterization of the C. vulgaris cell lines obtained following the stress treatmentsGrowth parametersThe cell lines recovered were subcultured in 50ml of Cildáñez stream water supplemented with the corresponding stressor (Phen, H2O2, or I) under the conditions described previously. Growth was estimated daily by monitoring absorbance at 680nm. Control treatments were added:
- 1.
LMPA-40 strain maintained in MS synthetic medium until the last subculture to the corresponding treatment (tolerance treatment, stressor+TT).
- 2.
LMPA-40 strain maintained in MS medium subcultured to the corresponding treatment for 6 subcultures before use (short acclimation, stressor+SA)
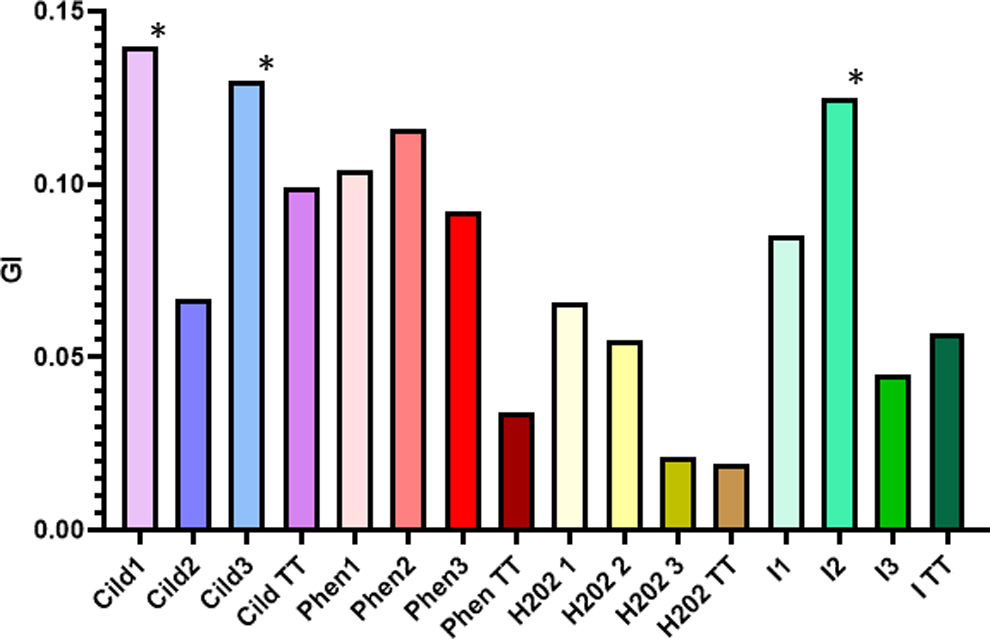
For each population, the relative growth rate (RGR) was estimated as the growth at day n with respect to the initial point (time 0) and growth index (GI) as the relation between RGR and the period estimated at day 4th with respect to the initial point (time 0).
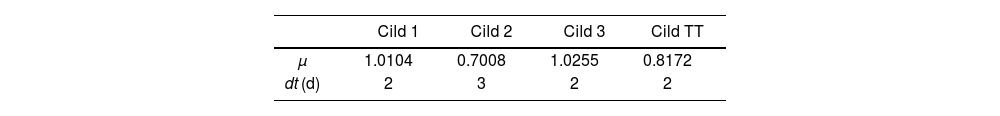
Stoichiometric parameters were determined using Fermentool software (https://fermentertool.com/) after 11 days of culture. Kinetic cell growth was estimated using the formula:
where x represents the biomass obtained at time (t), and μ is the specific growth rate. The duplication time (dt) was calculated as ln (2)/μ.Volumetric productivity (P) was also calculated as follows:
where x is the biomass obtained in a volume (vol.) in a unit of time (t).Comparison of the Cild 3 line with respect to TT and SA. Biochemical assaysAfter 6 months of maintaining the Cild 3 line in MS medium, biochemical assays were conducted culturing the cells in 30ml of water from the Cildáñez stream for 7 days under the previously described conditions. To ensure the independence of the samples, as many Erlenmeyers as points to be analyzed were processed.
Protein quantification was performed using the method proposed by Lowry et al.17. Algal biomass (1ml) was centrifuged at 10000g for 5min. The pellet was treated with 75μl of 1M NaOH in a water bath at 100°C for 1h. For the colorimetric reaction, 50μl of the sample diluted 1:10 was used. Absorbance at 750nm was measured in a spectrophotometer (Biotek μQuant™). Bovine serum albumin (BSA) was used for the standard curve.
Lipid quantification was performed by the sulfo-phospho-vanillin method according to Mishra et al.20 with modifications. Briefly, a mixture of 2ml of sulfuric acid and 0.1ml of the algal biomass (1ml) was centrifuged at 10000g for 5min, then heated for 10min at 100°C and cooled for 5min in an ice bath before the addition of 5ml of phospho-vanillin (vanillin 0.2mg/ml of 15% phosphoric acid). The reaction was shaken at 200rpm for 15min and absorbance at 530nm was recorded. Canola oil dissolved in chloroform (0–4mg lipids/ml) was used to prepare the standard curve.
For pigment quantification, the methodology described by Wegmann and Metzner34 was followed. Briefly, 3ml of biomass was centrifuged at 16100g for 5min. The pellet was resuspended in 1.5ml of distilled water and centrifuged again (16100g, 5min). The obtained pellet was frozen in liquid nitrogen, subsequently resuspended in 1.5ml of acetone, and incubated for 24h at 4°C before measuring the absorbance at 453, 644, and 663nm, using 90% acetone as blank.
Chlorophyll A, B, and carotenoids were quantified using the following formulas:
Statistical analysisPhysicochemical and microbiological analyses were performed at the beginning and at the end of the bioprocess (day 7). Analytical determinations were performed by triplicate.
The Student t-test was used to establish the significant difference between the response of two groups. The normal distribution of the data sets was confirmed through the Shapiro–Wilks test. The equality of variances was assessed using the F test with a significance threshold of 0.05. Moreover, results were evaluated by ANOVA with post-hoc Tukey’ s test29 for multiple comparisons, or by the Kruskal–Wallis test for non-normal variables, using Infostat software8.
Results and discussionALE and random mutagenesis assays on C. vulgarisThe stressor concentrations used for the experiments, chosen from preliminary assays (data not shown), allowed the algae to continue growing in subsequent subcultures. At higher stressor concentrations or longer irradiation times, subsequent subcultures of this algae line were not possible.
After 110 days under selective pressure, the adapted algal cell lines were isolated by removing microorganisms from Cildáñez stream water, eliminating their competitive impact. This step was included in the experimental design based on previous research, which showed that C. vulgaris strain LMPA-40 exhibited greater growth rates in autoclaved Lake Lugano water (near Cildáñez) than in non-autoclaved water, due to the competition for nutrients with the microbiota present in the water7.
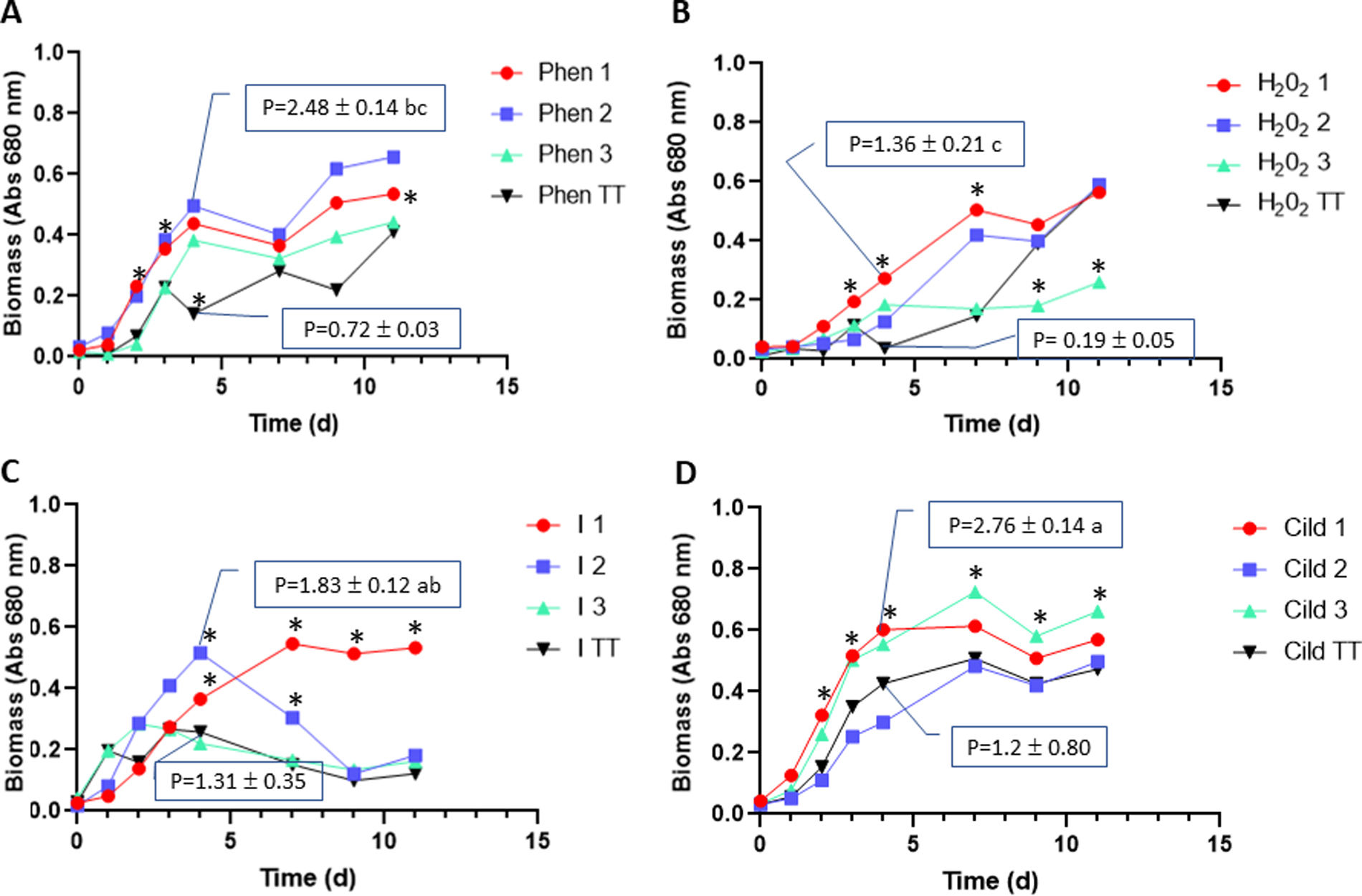
Cell lines obtained under Phen selection pressure showed a greater resistance capacity (p-value 0.002) than the Phen TT line when they were cultivated in Cildáñez water for 11 days (Fig. 1A). The biomass of the three Phen lines increased until day 4, when they entered the stationary phase. GI of the three Phen populations showed significant differences concerning Phen TT at 4 d (Fig. 2). Similar results were found by Wang et al.32 who reported a C. vulgaris strain resistant to 500mg/l of phenol after 31 culture cycles under selection pressure. Thus, the adapted strain was able to double the resistance to phenol compared to the original strain. In our case, the phenol resistance of adapted cells was threefold greater than that of the original strain. It should be noted that our experiments were conducted in Cildáñez water, which increases stress conditions.
Growth assays after ALE and random mutagenesis. C. vulgaris cell lines (1–3) purified after ALE, random mutagenesis or control line (TT) grown in Cildáñez water supplemented with Phen (160mg/l) (A), H2O2 (100mg/l) (B), I (UV-C for 12min) (C) or Cildáñez water without any contaminant (D). Asterisks show significant differences between treatments in a time at p<0.05. The maximum volumetric productivity (P) achieved, which estimates the amount of biomass generated per unit of culture in a unit of time, is reported in a square for the lines that showed the best results in the kinetics assays performed with the lines maintained in MS during 3 months after the ALE and mutagenesis assays and, also, for the same control line (TT).
Growth index for all C. vulgaris cell lines obtained from the ALE and random mutagenesis assays. C. vulgaris cell lines purified after ALE (1–3), ALE plus mutagenesis (I) or TT lines grown in Cildáñez water supplemented with Phen (160mg/l), H2O2 (100mg/l), I (UV-C for 12min) or Cildáñez water without any contaminant. The growth index was calculated on the 4th day of growth. Asterisks show significant differences between treatments at p<0.05.
The cell lines isolated and purified from H2O2 treatments showed no resistance to this agent during the first 3 days of culture with selective pressure (Figs. 1B and 2). For lines H2O2 1 and 2, the growth slope accelerates from day 3 to day 7 when the maximum biomass is reached (μ 0.760 and 0.920 resp.) with a dt of 3 d for line 1 and 2 d for line 2 and maximum GI at 4 d on culture. The H2O2 3 line did not overcome the stress and exhibited almost no growth. On the contrary, the H2O2 TT line overcame the stress and began to grow from day 7 to 11 (μ=0.9212), reaching the same final biomass as lines H2O2 1 and 3.
During the first three days of growth, none of the three cell lines isolated from the UV-C treatment differed from the I TT treatment. Only line I 1 showed tolerance to irradiation with significant differences compared to I 2, I 3 and I TT (Figs. 1C and 2) after day 7 (p-value 0.0007). Line I 2 exhibited a higher growth from day 2 to 4 and higher GI compared to the other treatments; however, growth decreased from day 4, reaching a similar final biomass to that of I TT and I 3. Thus, the curves for cell lines I 2 and I 3 are comparable to those obtained with the TT line, demonstrating that the adaptations observed at the end of the ALE plus the random mutagenesis assays were reversed during subculture in MS medium.
The GI allows to quickly visualize the cell lines that modified their growth after the applied treatments (Fig. 2). However, after comparing the results obtained with the same assay driven with yeast or bacteria, a low frequency of obtaining new cell lines was observed26. This difference, in the frequency of obtaining new cell lines, is usually attributed to the low growth rate and large genome size of green microalgae36.
In the Cild treatment, Cild TT and Cild 2 cell lines were the most affected (Figs. 1D and 2). Cell lines Cild 1 and 3 showed a similar behavior. While Cild 1 stopped growing on day 4, Cild 3 continued growing until day 7, reaching the maximum biomass at the end of the assay. These lines had the highest growth speed and GI with the lowest dt (Figs. 1D, 2 and Table 1). On day 4, the TT treatment showed the lowest GI demonstrating C. vulgaris susceptibility to all the stressors applied including Cildáñez water (p-value 0.0064) (Fig. 2 and Table 1). In Cild 1 and Cild 3, there was an adaptation process throughout the ALE trial that allowed them to reach higher growth rates compared to the TT treatment, as was also previously observed by other teams21,25.
Maximum volumetric productivity (P) achieved, which estimates the amount of biomass generated per unit of culture volume in a unit of time, is reported for the lines that showed the best results in the kinetics assays performed (Fig. 1 data in squares, lines Phen 2, H2O2 1, I 2, Cild 3 and their respective TT controls). Maximum P was achieved in the Cild 3 line in consonance with the previous results observed (Figs. 1 and 2 and Table 1) followed by Phen 2 and lastly, I 2 and H2O2 1. The low P observed in the control treatments (TT) showed the negative effect of stress in lines that were not adapted to grow in the presence of each stressor.
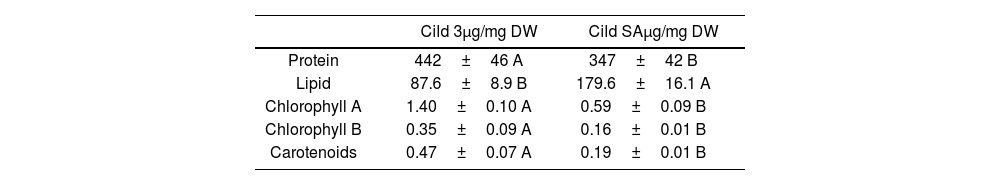
Biochemical characterization of C. vulgaris strains Cild 3 and Cild SAALE strategies under different stress conditions were shown to alter the algal metabolite content14,28,33. Given that Cild 3 showed the best results in the previous experiments (Figs. 1 and 2), the biochemical parameters of this strain after 7 days (the end of log phase for Cild 3 cell line) of culture in Cildáñez water were compared against the Cild SA control treatment.
The protein concentration of the Cild 3 line was 27% higher than the Cild SA line (Table 2). It is possible that the microalgae adapted to grow in contaminated water respond more quickly to stress by increasing the synthesis of defense proteins, as has been seen in Euglena gracilis exposed to cadmium stress that increased the levels of thio-rich proteins and proteins involved in the stress response15. A 10.5% increase in protein content was reported in the ALE experiments to improve high-temperature tolerance of Chlorella sorokiniana although their lipid content also increased by 20.95% and that of carotenoids by 81%28. All these metabolic changes in this strain led to the adoption of a multi-system synergistic mechanism to enhance antioxidant capacity, maintaining protein homeostasis, remodeling photosynthetic metabolism, and regulating the synthesis of heat-stress related metabolites, which enabled the strain to adapt to the high-temperature conditions28. On the other hand, other stresses had different effects, e.g., carbohydrates and protein content were significantly decreased while lipid content was 2.2-fold higher in the hypersaline conditions ALE evolved C. vulgaris strains compared with the wild-type31. Cild 3 showed half the lipid content of Cild SA (Table 2). It is widely described that lipid concentration in microalgae usually increases when growth is inhibited by stress18,28,33. The lower lipid concentration of Cild 3 line could be a consequence of its adaptation to the adverse conditions of the culture.
Biochemical parameters estimated in the Cild 3 cell line purified after the directed evolution experiments and in the control treatment (Cild SA) grown in Cildáñez water after 7 days of culture.
| Cild 3μg/mg DW | Cild SAμg/mg DW | |
|---|---|---|
| Protein | 442±46 A | 347±42 B |
| Lipid | 87.6±8.9 B | 179.6±16.1 A |
| Chlorophyll A | 1.40±0.10 A | 0.59±0.09 B |
| Chlorophyll B | 0.35±0.09 A | 0.16±0.01 B |
| Carotenoids | 0.47±0.07 A | 0.19±0.01 B |
Letters indicate significant differences (p<0.05).
Pigment concentration is another biochemical parameter usually used to assess tolerance to a contaminated medium, since in many cases they are inhibited by stress1,6,24. In our assay Chlorophyll A, Chlorophyll B and carotenoid concentration in Cild 3 were significantly higher than in Cild SA (Table 2). It is known that carotenoids play a role in stress tolerance in microalgae27,29. These metabolites are involved in the primary metabolism pathways, which could be important in future studies to elucidate the mechanism by which this resistance was achieved.
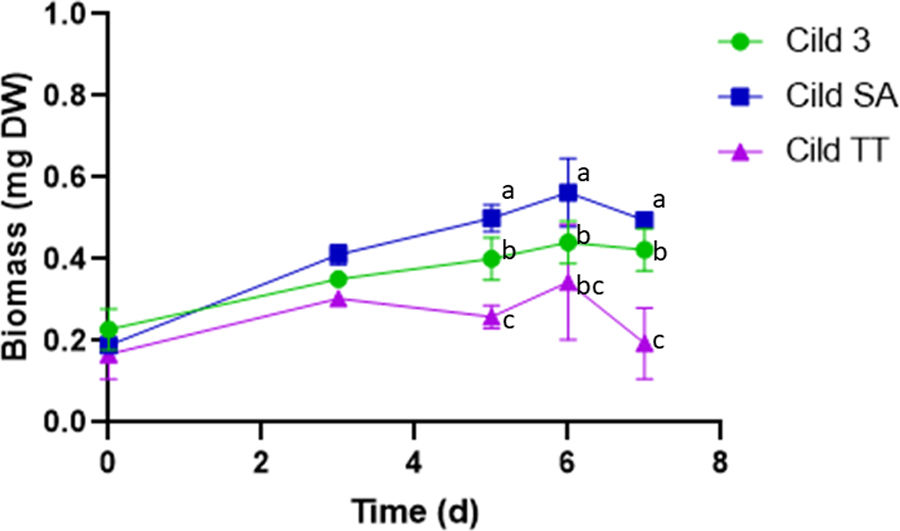
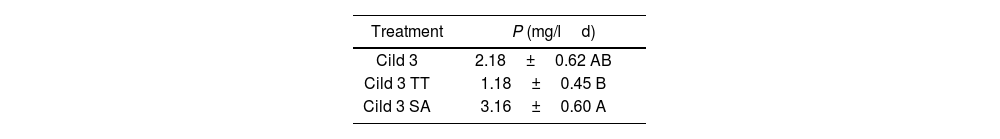
After 6 months of maintenance of the Cild 3 cell line in MS medium, the assay in Cildáñez water was repeated (Fig. 3 and Table 3). The results obtained were different from those observed in previous trials. The best results were obtained with Cild SA with high accumulation of biomass after 6 days of treatment with similar specific growth rate (μ=0.200) and dt (3 d) as Cild 3 (μ=0.199 and dt=3) and high P at the 4th day of treatment (Table 3). A short period of acclimatization to this water (Cild SA, the cell line maintained in MS medium subcultured onto the corresponding treatment for 6 subcultures before use), probably elicits a stress response, achieving a higher biomass than Cild 3 and Cild TT treatments (same Fig and Table). These results are in line with previous observations7,13,19. It is important to highlight that the value in the Cild 3 cell line after 6 months cultured in MS medium without stress (Table 3, P=2.18mg/mld) was significantly lower (p-value 0.046) than that observed for the Cild 3 cell lines purified after ALE (Fig. 1, P=2.76mg/mld).
C. vulgaris biomass (abs 680nm) of the Cild lines grown in Cildáñez water for 7 days after 6 months of maintenance in MS media without stress. Cild 3 purified cell line after ALE assay; Cild TT, original strain (LMPA-40 strain) maintained in MS synthetic medium until the last subculture to the Cildáñez water; Cild SA, original strain (LMPA-40 strain) maintained in Cildáñez water during the last six subcultures. Letters indicate significant difference at p<0.05.
Volumetric productivity (mg biomass/unit of volumeday) at day 4, obtained from C. vulgaris cell line Cild 3 after 6 months of culture in MS media without stress (see Fig. 3), grown in Cildáñez water for 7 days.
| Treatment | P (mg/ld) |
|---|---|
| Cild 3 | 2.18±0.62 AB |
| Cild 3 TT | 1.18±0.45 B |
| Cild 3 SA | 3.16±0.60 A |
Capital letters indicate significant differences (p<0.05) of the Cild 3 cell line or control Cild 3 lines (TT o SA) after 6 months of maintenance in MS (p<0.05).
This result shows that the acclimatization period was more significant than the mutations that could have occurred in the Cild 3 cell lines. Considering that the Chlorella genus evolved on a primitive earth with many different environmental conditions, such as a different atmosphere composition10, it was able to overcome all the changes occurring from the beginning to the present day, demonstrating the genome plasticity which allows the ability to grow in extreme conditions5,9. This genome plasticity would explain the cell adaptation after successive cultures of algal cells in the presence of a stressor. However, unlike the cells obtained after ALE adaptation to contaminated water, which likely adapted through genome mutations, the algae managed to grow in the short acclimatization period by responding to stress with an increase in lipid content, showing an acute response.
ConclusionsThe current study focused on the exposure and adaptation of stressors, resulting in the generation of improved C. vulgaris cell lines that exhibited enhanced resistance to contamination. The isolated and characterized Cild cell line that displayed tolerance to Cildáñez water exhibited significant differences in various biochemical parameters compared to the SA line that could be related to the acclimatization process. Directed evolution and its combination with random mutagenesis allowed to obtain cell lines with greater growth than the TT line in the Cild, Phen, and I treatments. This indicates their possible utility for bioremediation processes. However, not all the cell lines obtained after ALE could adapt enough to overcome the stress caused by contaminated water. This suggests that the process is quite random and depends on the stressor used. Additionally, the acquired tolerance could be lost with storage time. The results obtained raise concerns regarding the feasibility of investing resources in the acquisition of novel cell lines.
FundingThis research was supported by Cyted-Renuwal320RT0005.
Authors’ contributionsJ.M.N.L., M.D.G. and P.L.M.; methodology, U.S. and A.G.T.; validation, U.S., A.G.T. and J.G.S.N.; formal analysis, J.M.N.L., M.D.G. and P.L.M.; investigation, U.S.; resources, J.M.N.L. and P.L.M.; data curation, US; writing – original draft preparation, J.M.N.L., M.D.G. and P.L.M.; writing – review and editing, J.M.N.L., M.D.G. and P.L.M.; funding acquisition, J.M.N.L. and P.L.M. All authors read and approved the submitted version.
Conflict of interestThe authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We are grateful to the Iberoamerican network 320RT0005-Renuwal-Cyted. We would like to thank Dr. S. Ochatt for critical reading of this manuscript.