Coagulase-positive staphylococci (CoPS) are opportunistic pathogens carrying various mechanisms of resistance that have a large number of virulence factors, and whose ability to induce illness is associated with the host. This study aimed to investigate the presence of environmental coagulase-positive staphylococci, their susceptibility profile, clonal relationship and ability to form biofilm. The 16S rRNA genes from CoPS isolates were analyzed, and their antibiotic susceptibility was evaluated using the agar dilution method in accordance with Clinical and Laboratory Standards Institute guidelines. The clonal profile was obtained by pulsed-field gel electrophoresis (PFGE) and biofilm formation was measured by a crystal violet retention assay. A total of 72 Staphylococcus spp. strains were isolated from air, metal surfaces, and nostrils from humans, dogs, cats, and birds. Three species were identified: Staphylococcus aureus (17%), Staphylococcus intermedius (63%), and Staphylococcus pseudintermedius (21%). Ninety three percent (93%) of the strains were resistant to at least one of 13 tested antibiotics. S. pseudintermedius strains were the only resistant ones to methicillin while most of these isolates were multidrug-resistant, had significantly higher ability to form biofilm and PFGE grouped into seven different patterns, without showing clonal dispersion among animals and environmental isolates. This study suggests that dogs, cat, and air are environmental sources potentially carrying multidrug-resistant S. pseudintermedius, which survives in different environments through biofilm formation and multidrug resistance, characteristics that can be transmitted horizontally to other bacteria and exacerbate the problem of antibiotic resistance in humans.
Los estafilococos coagulasa-positiva (CoPS) son patógenos oportunistas, portan varios mecanismos de resistencia, tienen un gran número de factores de virulencia y su capacidad para inducir la enfermedad está asociada con el hospedero. El objetivo de este estudio fue investigar la presencia de CoPS en el medio ambiente, su perfil de sensibilidad a los antibióticos, su relación clonal y su capacidad para formar biopelícula. De los aislamientos de CoPS se analizaron los genes 16S ARNr y se evaluó la sensibilidad a los antibióticos mediante el método de dilución en agar según el CLSI. El perfil clonal se obtuvo por electroforesis en gel de campo pulsado (PFGE) y la formación de biopelícula se analizó por retención de cristal violeta. Se aislaron 72 cepas de Staphylococcus spp. a partir de aire, superficies metálicas y narinas de humanos, perros, gatos y aves. Se identificaron tres especies: Staphylococcus aureus (17%), Staphylococcus intermedius (62%) y Staphylococcus pseudintermedius (21%). El 93% de las cepas fueron resistentes al menos a uno de 13 antibióticos probados. Los aislamientos de S. pseudintermedius fueron los únicos resistentes a meticilina y la mayoría fueron resistentes a múltiples fármcos, tuvieron una capacidad significativamente mayor para producir biopelícula y la PFGE los agrupó en 7 diferentes patrones, sin mostrar dispersión clonal entre los aislamientos de animales y de medio ambiente. Este estudio sugiere que los perros, los gatos y el aire son fuentes ambientales potencialmente portadoras de S. pseudintermedius resistente a múltiples antibióticos. Este agente sobrevive en diferentes entornos en virtud de la formación de biopelículas y la resistencia a múltiples antibióticos, características que pueden transmitirse horizontalmente a otras bacterias y, por ende, exacerbar el problema de la resistencia a los antibióticos en humanos.
Staphylococci are catalase-positive Gram-positive cocci. The different species can be distinguished by their ability to ferment sugars and produce coagulase. Seven species of coagulase-positive staphylococci (CoPS) have been identified: Staphylococcus aureus, Staphylococcus intermedius, Staphylococcus schleiferi subsp. coagulans, Staphylococcus hyicus, Staphylococcus lutrae, Staphylococcus delphini and Staphylococcus pseudintermedius7. CoPS commonly colonize the skin and mucous membranes; furthermore, staphylococci have the ability to survive in almost any environment. CoPS are opportunistic pathogens; they have a large number of virulence factors, and their ability to induce illness is usually associated with the host. CoPS carry various mechanisms of resistance. Particularly, S. aureus became methicillin-resistant by acquiring a genomic island of resistance known as chromosomal cassette mec (SCCmec I-VII), and is a variable genetic element. The island is present constitutively in the orfX gene, and depending on the type, has a specific recombinase ccr, which allows to carry other resistance genes harbored in small plasmids or transposons12.
The epidemiology of staphylococci has changed in recent years, as they can cause nosocomial and community infections, and the importance of S. aureus has increased because it can cause many pathological conditions ranging from simple skin infections to invasive processes such as pneumonia and osteomyelitis. Moreover, Staphylococcus epidermidis is considered a harmless commensal bacterium of the human skin, even an accidental pathogen16. However, at present, this bacterium is recognized as an important human pathogen and is one of the main causes associated with medical devices such as peripheral or central intravenous catheter-related infections. It also causes keratitis and endophthalmitis, contamination of contact lenses, urinary catheter infections, bacteremia, mediastinitis and other infections. Both species are reported to have high rates of resistance to methicillin, and there is an increasing number of reports on their reduced susceptibility to vancomycin5,20,21.
It is known that staphylococcal species exhibit host specificity, and the species of clinical CoPS specimens differ from those isolated from animals, which also differ among host species. For example, the predominant species in ruminants, pigs, dogs and pigeons are S. aureus, S. hyicus, S. pseudintermedius and S. intermedius respectively6,23. However, recent studies consider S. pseudintermedius as an emerging zoonotic agent17,29.
Although there is great clonal variability among staphylococci, it is not understood why some Staphylococcus clones have greater dissemination or why some species are more prevalent than others. In addition, many of these disseminated species that are distributed throughout the world may even replace native clones, although only some of the disseminated species have been reported to cause infection. In the U.S.A., it has been observed that the USA300 and USA400 clones belonging to sequences ST8 and ST1, respectively, cause most community-acquired S. aureus infections, and the USA100 clone of S. aureus is disseminated in hospital environments26. In addition, pseudintermedius can be spread in humans and their pets. In 2006, the first case of infection by this microorganism in humans was reported27, and from 2010 to 2012, S. pseudintermedius was responsible for causing an outbreak in a veterinary hospital for dogs and cats of difficult control28,29. Furthermore, the spread of clones ST71 in Europe and ST68 in America has been observed among pets8,24.
This study aimed to investigate the presence of environmental coagulase-positive staphylococci as carriers of resistance factors, their clonal relationship and ability to form biofilm.
MethodsBacterial isolatesAirborne bacteria were collected from 10 different areas of the Veterinary Teaching Hospital (Mexico city, D.F. Mexico) using a two-stage Andersen sampler (Graseby Andersen, Atlanta, GA), with a constant air flow rate of 28l/min for 15min. Samplers were loaded onto Petri dishes containing blood agar and trypticase soy agar (Difco Laboratories, Detroit, MI). A total of 10 surface samples from stainless steel tables within 5cm2 areas were collected using a swab technique. The animal samples were taken from the nostrils (from 20 dogs, 13 cats and 16 birds), were collected using a swab technique and cultured on the same media; 10 human nasal exudate samples were also collected. All the agar plates were incubated for 24–48h at 37°C. Typical colonies of staphylococci were transferred onto mannitol salt agar selective medium (Difco Laboratories). The coagulase test was performed with rabbit plasma on mannitol-positive organisms. Bacterial strains (coagulase- and mannitol-positive) were identified by 16S rRNA sequencing.
Human, animal, air and surface samples were collected at the same time and from the same space. (Site: hospital for dogs, cats and birds.)
PCR amplification of 16S rDNA genesPartial 16S rDNA gene sequences were amplified by PCR using universal primers 27F and 1492R. Primer 27F was 5′-AGA GTT TGA TCM TGG CTC AG-3′, and primer 1429R was 5′-TAC GGY TAC CTT GTT ACG ACT T-3′. The PCR reaction mixture included 2μl of bacterial DNA, 35.4μl of ddH2O, 5μl of 10× buffer, 1.5μl of MgCl2 (1.5mM), 1μl of dNTPs (10mM), 0.1μl of Taq (20μl), and 2.5μl each primer (10μmol) in a final reaction volume of 50μl. Amplifications were performed as follows: 94°C for 1min; 94°C for 1min, 56°C for 30s, 72°C for 1min 30s (35 cycles); 72°C for 5min; and then a hold at 4°C. PCR products were examined for size and yield using 1.2% (w/v) agarose gels in TAE buffer. After successful amplification, the obtained products were sequenced using a PRISM 3730 automated sequencer (Applied Biosystem Inc.).
Sequence analysisDNA sequences were edited and assembled using the SeqMan and Edit Seq programs (DNA Star, Laser Gene 6, USA). Sequence similarity analysis was performed using the BLAST program (http://www.ncbi.nlm.nih.gov/BLAST).
Antimicrobial susceptibilityAntimicrobial susceptibility of isolated Staphylococcus strains was tested using Vitek I GPS-119 cards (bioMérieux, Inc., Durham, NC). Resistance was verified by Minimum Inhibitory Concentration (MIC), employing the agar dilution method as described by the Clinical and Laboratory Standards Institute guidelines (CLSI 2014)4. The antibiotics used were: penicillin-G (MP Biomedicals), oxacillin (Sigma–Aldrich, St. Louis, MO), Ampicillin (MP Biomedicals, Solon, OH), clindamycin (Sigma–Aldrich), erythromycin (MP Biomedical), gentamicin (MP Biomedicals), levofloxacin (Sigma–Aldrich), moxifloxacin (Sigma–Aldrich), tetracycline (Sigma–Aldrich), trimethropim (MP Biomedicals), sulfametoxazole (MP Biomedicals), and vancomycin (MP biomedical). S. aureus reference strain ATCC® 29213 (American Type Culture Collection, Manassas, VA, USA) was used. Isolates exhibiting oxacillin resistance were also confirmed by the presence of the mecA gene using previously described primers and terms30.
Assay for biofilm formationQuantification of biofilm formation was performed using 96-well polystyrene microtiter plates (Costar flat-bottom plates with lids) in accordance with Stepanovic's method with slight modification25. Briefly, bacterial cells were grown overnight in TSB, either from a single colony grown on TSB agar or from a glycerol stock kept at −70°C. Cells were diluted 1:100 in 200μl of TSB or TSB+glucose (1%). Each bacterial strain was assayed in four replicate wells. Microtiter plates were incubated at 37°C. After 24h of incubation, total cell growth was measured based on optical density (OD) at 570nm using a Bio-Tek Elx808 microplate reader with the Kc4 software. Planktonic cells were discarded, and the plate was treated with 200μl volumes of the following reagents: after rinsing three times with PBS 1×, the remaining biofilm was fixed with methanol (100%), stained with crystal violet (2%), and rinsed with water three times. The dye was then solubilized with acetic acid (33%). Finally, the OD570nm was determined using a microplate reader. The amount of biofilm formed is reported as the ratio of OD570nm/OD620nm values, which corresponds to a simplified expression of the ratio used in previous studies15.
Pulsed field gel electrophoresis (PFGE)PFGE analysis was performed for the S. pseudintermedius and S. intermedius isolates. The procedures and buffers used for the preparation of chromosomal DNA, macro-restriction of DNA, and PFGE were modified from an earlier report14. Briefly: digestion was performed in a volume of 200μl with 1× enzyme buffer and 25U SmaI were incubated at 25°C overnight. A 1% (wt/vol) agarose gel was prepared in 0.5× TBE buffer (AMRESCO, Solon, OH). PFGE was performed using a multistate program and a CHEF Mapper system (Bio-Rad, Hercules, CA). The running parameters were as follows: 200V (6V/cm); temperature 14°C; block one: initial switch 2s, final switch 7s, 10h duration; block two: initial switch 8s, final switch 45s for 14h. After the electrophoresis run was completed, the gel was stained in a 1.5μg/ml ethidium bromide solution (AMRESCO X328, 10mg/ml; Amresco, Inc., Solon, OH) for 20min in a covered container and destained in fresh distilled water for 45min. A Lambda DNA (New England Biolabs) was used as a molecular size standard.
Statistical analysisThe Statistics Package for the Social Sciences program (SPSS, version 10) was used for the parametric Student's t-test. Categorical variables were described as percentages, and median, minimum and maximum values were used for continuous quantitative variables.
PFGE data analysis was performed by considering the presence or absence of specific bands to obtain an estimate of similarity for each pair of isolates. Gel fingerprint patterns were analyzed using BioNumerics version 6.0 (Applied Maths). After background subtraction and gel normalization, fingerprint patterns were subjected to typing based on banding similarity and dissimilarity using the Dice similarity coefficient and the unweighted pair group method with arithmetic mean (UPGMA) clustering. The relationship was supported by the cophenetic correlation coefficient using Mantel and a bootstrap test with 10000 randomizations22. Multivariate statistical methods were performed using the NTSYS-PC program (version 2.0; Exeter Software).13
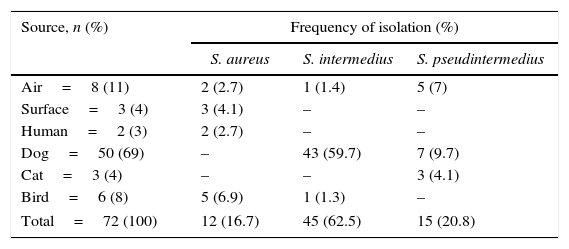
ResultsIsolation and identification of Staphylococcus spp.A total of 72 CoPS strains were isolated and identified. All isolates were confirmed by sequencing the ITS identification 16S gene. Fifty (50) CoPS strains were isolated from 10 of 20 dogs; 3 CoPS were isolated from one cat; 6 CoPS were isolated from two birds; two CoPS were isolated from one human, and 8 and 3 CoPS were isolated from the air and surfaces, respectively. Table 1 shows the frequencies of the observed species; the predominant species were S. intermedius (45/72), S. pseudintermedius (15/72) and S. aureus (12/72). The obtained S. aureus strains were not isolated from dogs or cats, and there were no differences between isolates of S. aureus and S. pseudintermedius (12 and 15, respectively). Three different species of CoPS (S. pseudintermedius, S. aureus and S. intermedius) were isolated from the air. Only S. aureus was isolated from the human patient and from the surfaces.
Frequency of coagulase-positive Staphylococci isolated from various sources in a veterinary school hospital in México
| Source, n (%) | Frequency of isolation (%) | ||
|---|---|---|---|
| S. aureus | S. intermedius | S. pseudintermedius | |
| Air=8 (11) | 2 (2.7) | 1 (1.4) | 5 (7) |
| Surface=3 (4) | 3 (4.1) | – | – |
| Human=2 (3) | 2 (2.7) | – | – |
| Dog=50 (69) | – | 43 (59.7) | 7 (9.7) |
| Cat=3 (4) | – | – | 3 (4.1) |
| Bird=6 (8) | 5 (6.9) | 1 (1.3) | – |
| Total=72 (100) | 12 (16.7) | 45 (62.5) | 15 (20.8) |
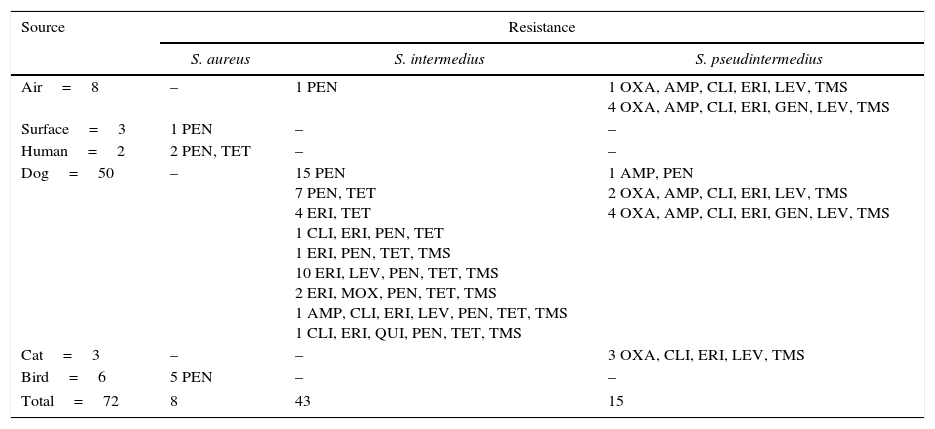
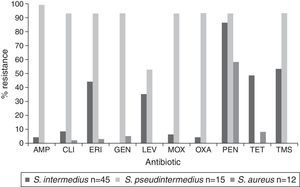
The observed antimicrobial susceptibility patterns are presented in Table 2. A total of 92% (66/72) of the strains of staphylococci showed resistance to at least one antibiotic, and 30% were Staphylococcus resistant to three or more different antibiotic families (MDR). Resistance was observed to penicillin, erythromycin, sulfamethoxazole/trimethoprim, tetracycline and levofloxacin (86, 47, 40, 40 and 39%, respectively). Only 14% (14/72) were resistant to oxacillin and they carried the mecA gene. S. intermedius was the most commonly isolated species and exhibited multidrug resistance in 16 cases, followed by S. pseudintermedius with 14 cases. None of the S. aureus isolates showed multidrug resistance, and it was the only species that was sensitive to all tested antibiotics in 4 of the 12 isolates. Figure 1 shows that the S. pseudintermedius species was resistant to most of the tested antibiotics, and all isolates showed resistance to ampicillin.
Antibiotic resistance patterns among coagulase-positive Staphylococci isolated from various sources in a veterinary school hospital in México
| Source | Resistance | ||
|---|---|---|---|
| S. aureus | S. intermedius | S. pseudintermedius | |
| Air=8 | – | 1 PEN | 1 OXA, AMP, CLI, ERI, LEV, TMS 4 OXA, AMP, CLI, ERI, GEN, LEV, TMS |
| Surface=3 | 1 PEN | – | – |
| Human=2 | 2 PEN, TET | – | – |
| Dog=50 | – | 15 PEN 7 PEN, TET 4 ERI, TET 1 CLI, ERI, PEN, TET 1 ERI, PEN, TET, TMS 10 ERI, LEV, PEN, TET, TMS 2 ERI, MOX, PEN, TET, TMS 1 AMP, CLI, ERI, LEV, PEN, TET, TMS 1 CLI, ERI, QUI, PEN, TET, TMS | 1 AMP, PEN 2 OXA, AMP, CLI, ERI, LEV, TMS 4 OXA, AMP, CLI, ERI, GEN, LEV, TMS |
| Cat=3 | – | – | 3 OXA, CLI, ERI, LEV, TMS |
| Bird=6 | 5 PEN | – | – |
| Total=72 | 8 | 43 | 15 |
Ampicillin (AMP), clindamycin (CLI), erythromycin (ERI), gentamicin (GEN), levofloxacin (LEV), moxifloxacin (MOX), oxacillin (OXA), penicillin-G (PEN), quinolone (QUI), tetracycline (TET), trimethroprim-sulfametoxazole (TMS).
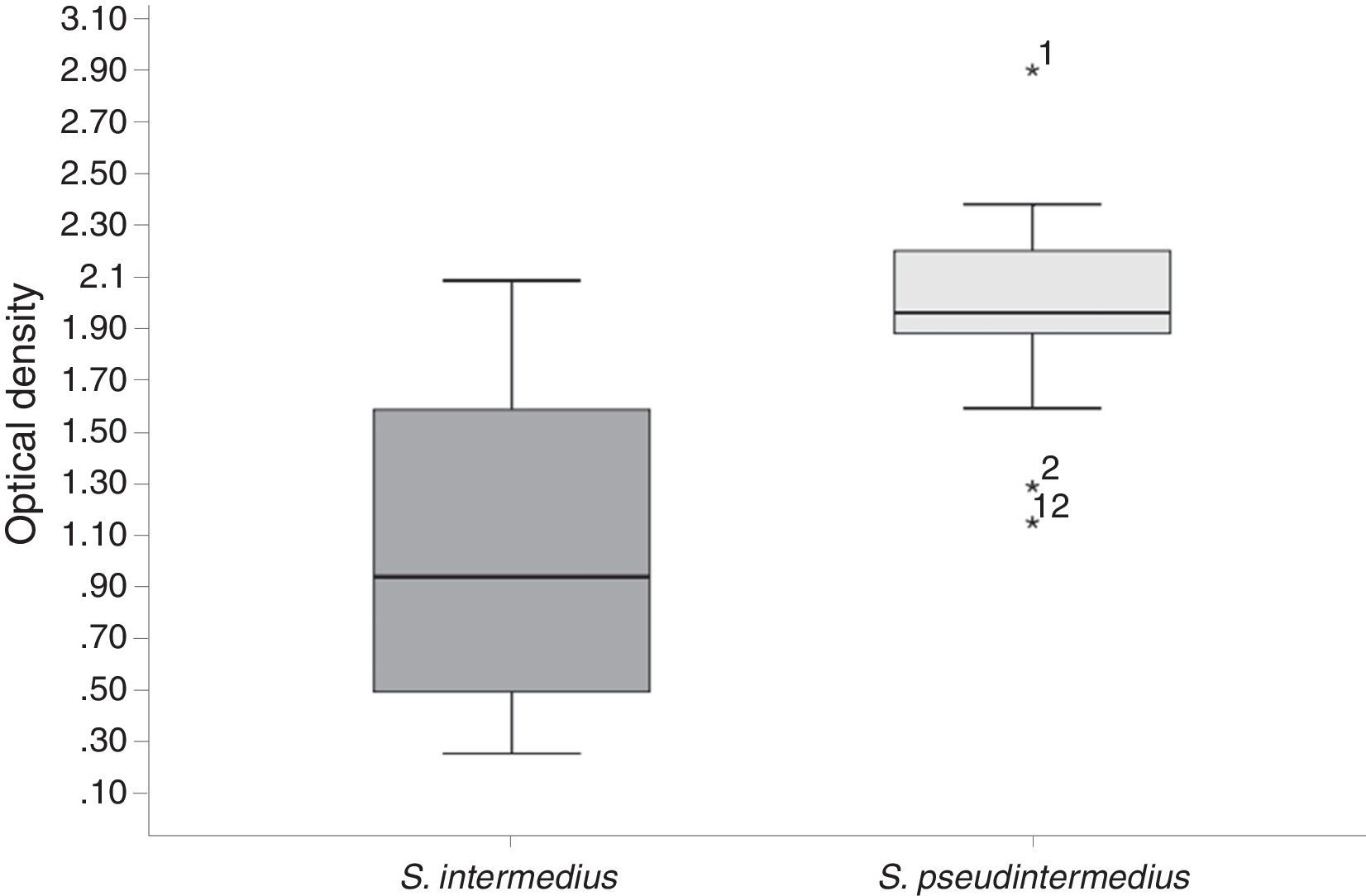
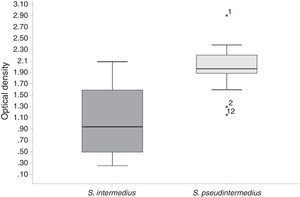
Biofilm formation was assessed after a 24h incubation period, and average OD570nm values were obtained (Figure 2). All of the strains in this study (CoPS) formed a biofilm (OD570nm <0.120 is considered not adherent, >0.240 is considered strongly adherent, and >0.120 to <0.2340 is considered adherent). E. coli strain BW25113 was used as a negative control. The S. pseudintermedius strains formed the greatest amount of biofilm and showed a statistically significant difference in biofilm-forming abilities compared with the other CoPS (S. intermedius and S. aureus). There was no statistically significant difference in biofilm-forming abilities among the S. intermedius and S. aureus isolates.
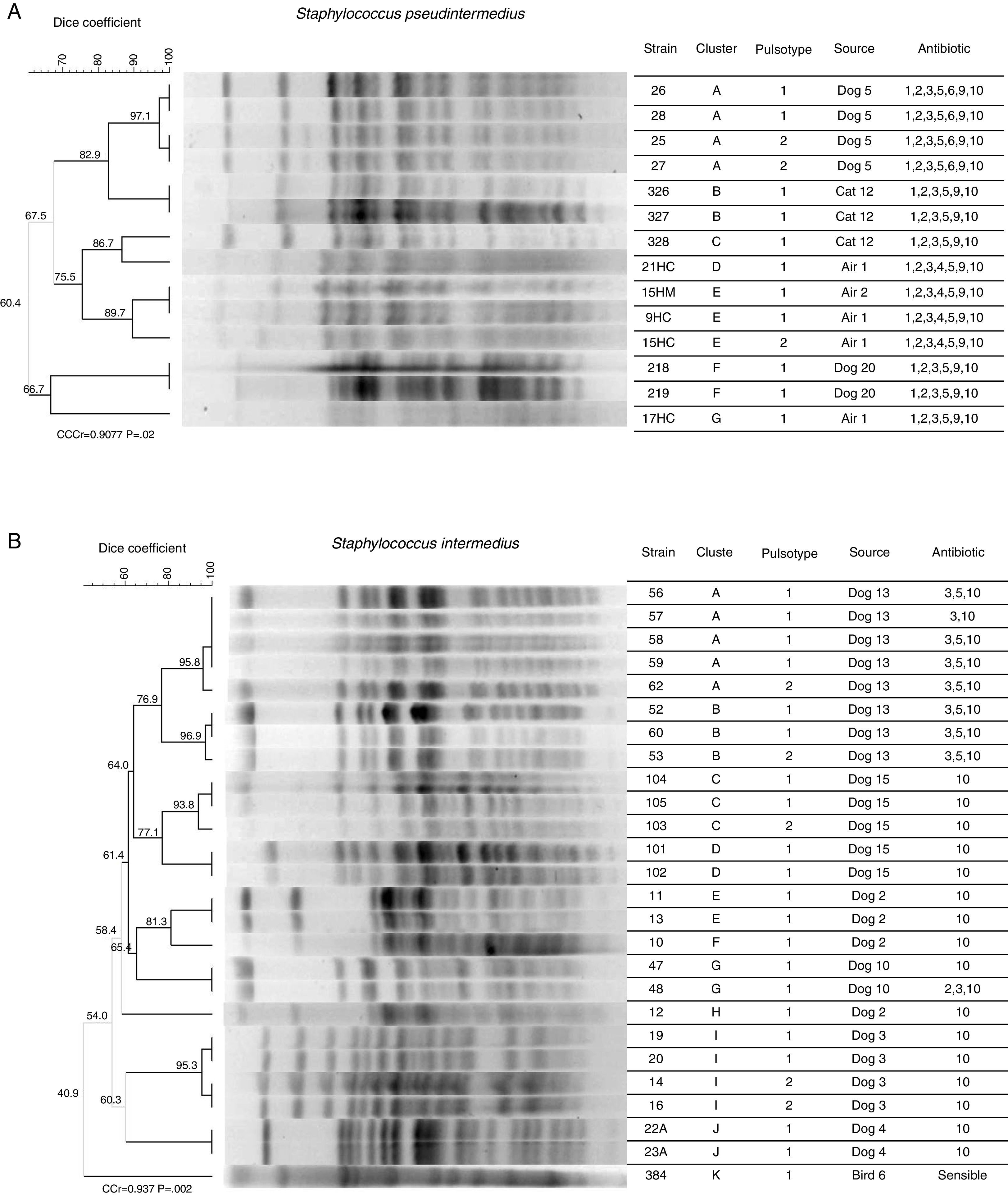
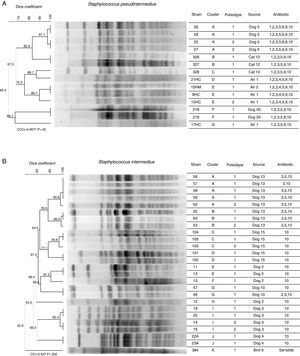
Pulsed field gel electrophoresis (PFGE)Figure 3 shows: (A) the clonal profile of S. pseudintermedius strains and (B) the clonal profile S. intermedius, the Dice similarity coefficient ranges were from 97.1 to 60.4% and 40.9 to 96.9% respectively. The banding patterns produced in S. pseudintermedius isolates from dogs and cats differed in four bands, meaning that there is a relationship between them. We observed that the antimicrobial susceptibility was also shared and only differed with respect to aminoglycoside (gentamicin). Two clusters with the same pattern of susceptibility were observed in S. intermedius isolates from the dog sample number 13. Finally, the PFGE patterns analyzed were grouped into 7 and 12 different patterns for S. pseudintermedius and S. intermedius strains, respectively.
Dendrogram analysis of PFGE patterns of S. pseudintermedius (A) and S. intermedius (B). Clonal profile analysis was conducted using the Dice similarity coefficient in association with the UPGMA algorithm as the grouping method. Dendrogram was evaluated by obtaining the cophenetic correlation coefficient with the Mantel test, which yielded an r-value (CCCr). Bootstrap values are given at the node.
Several studies have reported the circulation of CoPS between pets and their owners9,27. S. aureus is the species having the greatest impact on human health; however, other CoPS species may have major impact, such as the S. intermedius group, including S. intermedius, S. pseudintermedius, and S. delphini, which are closely related7. Generally, S. intermedius was considered to be the predominant staphylococci in dogs; however, recent evidence has shown that all or most isolates from dogs and cats previously identified as S. intermedius are actually S. pseudintermedius1. In this study, 16S rDNA gene sequencing was performed to identify and differentiate between species due to the problems arising from the use of conventional biochemical tests that have led to incorrectly reporting all isolates as S. aureus. Similar results were observed in a study in humans with dog bites, where S. pseudintermedius infections were incorrectly diagnosed as S. aureus3.
S. pseudintermedius and S. intermedius are opportunistic pathogens reported in the skin of dogs and have also been occasionally reported in serious zoonotic infections in humans11. Generally, pyoderma or skin infections are common elements in the medical practice in dogs and cats. In this work, S. pseudintermedius and S. intermedius were isolated from dogs, cats, birds and the air but not from humans. This demonstrates that S. intermedius and S. pseudintermedius are members of the normal flora of cats and dogs, which is the reason why these bacteria are commonly reported in a large number of clinical conditions in animals2.
Throughout the world, there have been increasing reports of the emergence of bacteria resistant to multiple antibiotics. In medical practice in humans, regulatory bodies have been set up regarding the use of antibiotics to control or reduce this problem. However, in agriculture and livestock, there are no regulations. The lack of regulation of broad-spectrum antibiotics in the veterinary clinic adds selection pressure on the normal flora bacteria, and allows them to become resistant to multiple antibiotics. The results obtained in this study confirmed the aforementioned findings: it was observed that different strains identified as S. pseudintermedius were resistant to multiple antibiotics in 90% of the study population. By contrast, S. aureus isolates were sensitive to multiple antibiotics in more than 90% of the study population. Surprisingly, strains of S. pseudintermedius showed 100% sensitivity to tetracycline, and S. aureus was 58% resistant to penicillin. In one study, S. pseudintermedius isolated from various diseases in cats showed greater than 90% resistance to the tested antibiotics, including tetracycline10. Additionally, the clonal profile analysis by PFGE is discriminative and very sensitive to the existing microvariation in a collection of strains; meanwhile, the type sequences are designed for tracking clones or clonal lines of bacterial populations. Data reported by Vigo et al., 201528, showed the great variability of a collection of strains of S. pseudintermedius isolated from infectious processes in dogs. They observed 27 different clonal types from 28 isolates. For our part, we noted that each animal or surface analyzed has its own clone of S. pseudintermedius or S. intermedius, and the same animal could be colonized by different clones. We also observed similarity coefficients over 80%, between cat and dog or cat and surface, presupposing a relationship between them. However, a limitation of this work is not knowing the type sequence of our strains, which would compare with those reported in other parts of the world. An example of this, is the work by Perreten et al., 201010,18, who reported two geographically distant scattered clones of S. pseudintermedius: clone ST71-J-t02-II-III in Europe and clone ST68-C-t06-V in North America. Both clones were isolated from various clinical conditions, including healthy animals. The ST71 clone was reported as a high biofilm producer and multidrug-resistant9,17, features also shown by our S. pseudintemedius strains.
These characteristics are relevant and not present in the S. intermedius strains in this work. Biofilm formation is a very important virulence factor and it is a highly variable feature among Staphylococcus species. However, biofilm formation ability has not been fully characterized in S. pseudintermedius. Studies performed by Zhou et al., 201331 and Pinheiro et al., 201419 showed an association between biofilm and the presence of the mecA gene and the icaADBC operon and our strains of S. pseudintemedius were the only ones where the mecA gene was identified. In addition, multidrug resistance is also associated with biofilm, because it has been recognized that antibiotics have limited diffusion in it, acting only on the bacterial surface, and further, antibiotics can react with other components of the biofilm matrix.
The main objective of this study was to identify possible sources of environmental contamination by opportunistic pathogenic bacteria in humans or their pets. It is also necessary to consider that multidrug-resistant microorganisms, such as S. pseudintermedius, survive in different environments through biofilm formation and multidrug resistance, characteristics that can be transmitted horizontally to other bacteria and exacerbate the problem of antibiotic resistance in humans.
Ethical disclosuresProtection of human and animal subjectsThe authors declare that no experiments were performed on humans or animals for this study.
Confidentiality of dataThe authors declare that no patient data appear in this article.
Right to privacy and informed consentThe authors declare that no patient data appear in this article.
Authors’ contributionsES, LM, ME participated in the study design, conducted the processing and analysis of samples in the laboratory, analyzed the data, and helped to draft the manuscript. ALO, RO contributed to manuscript writing. NVG and IR contributed to the conceptualization of the study and manuscript writing. All authors read and approved the final manuscript.
Conflict of interestThe authors declare that they have no conflicts of interest.
The authors thank Juan Carlos Vigueras Galindo for review of the manuscript. This work was supported by Federal Resources (HIM/2013/009 SSA 1081) from SSA.